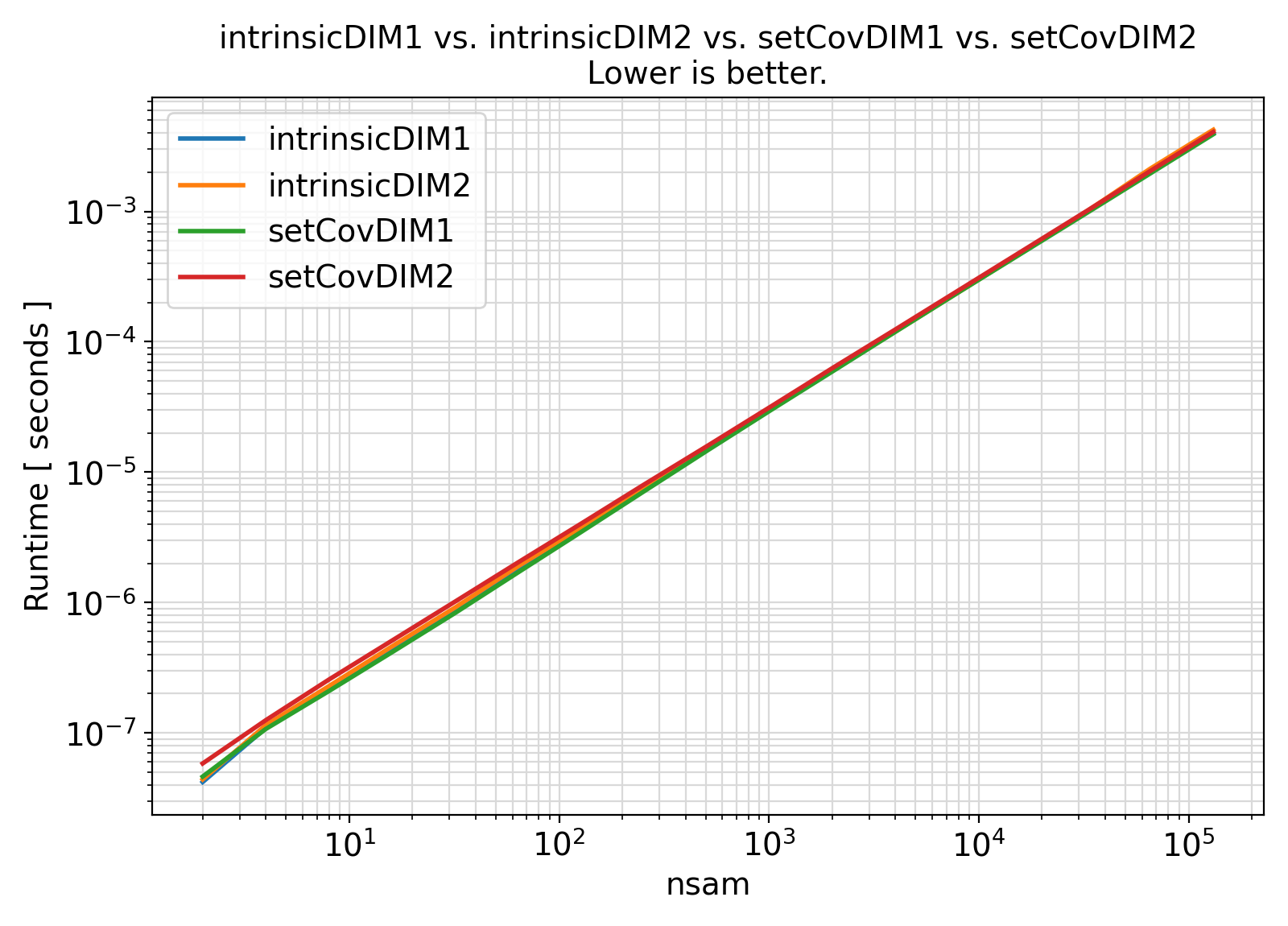
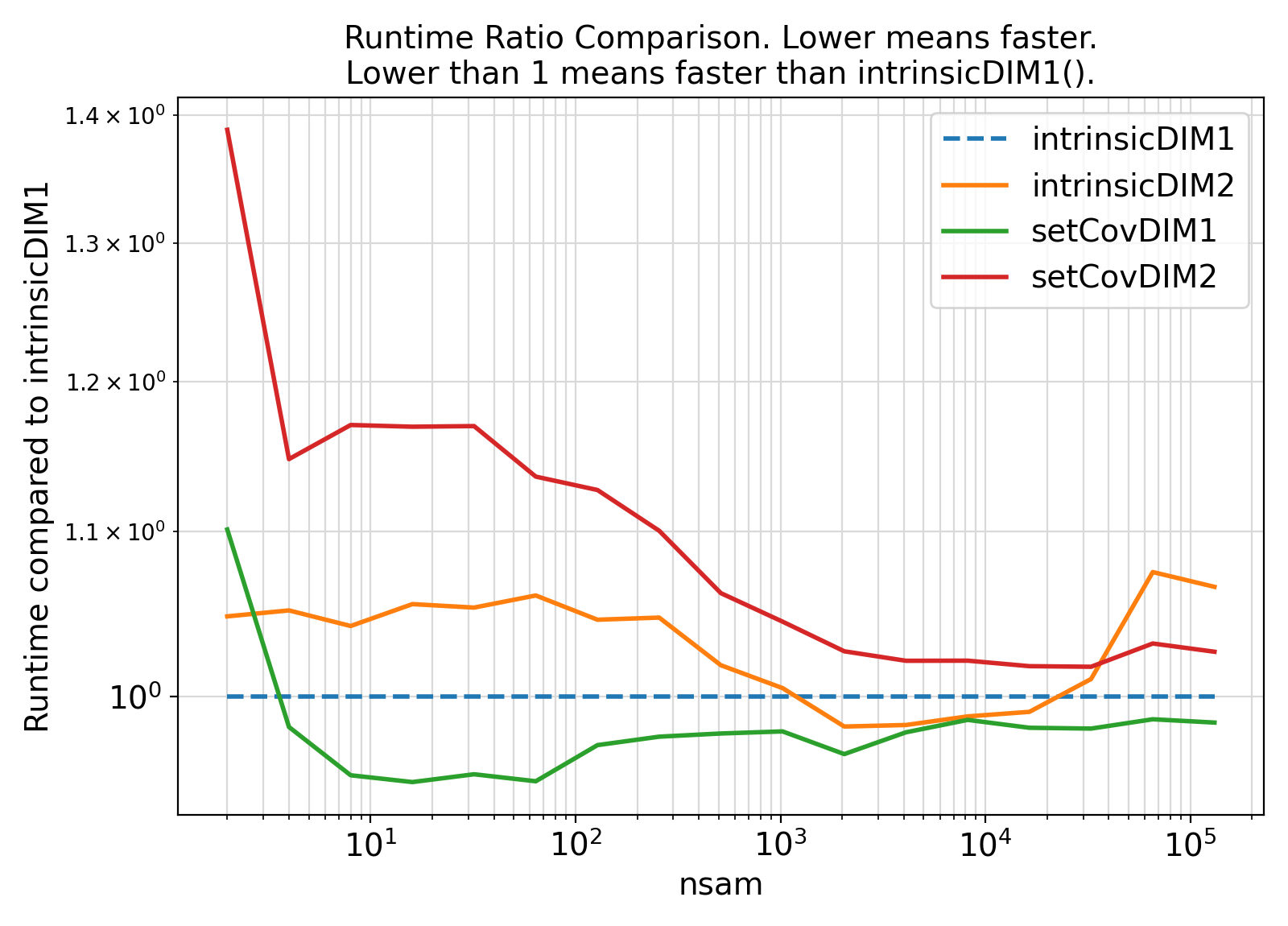
19 integer(IK) :: itry, ntry
= 10
20 type(display_type) :: disp
21 character(:),
allocatable :: format
25 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
26 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
27 call disp%show(
"!Convert correlation matrix and standard deviation to covariance matrix.")
28 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
29 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
37 real(TKG),
allocatable :: cov(:,:), cor(:,:), std(:)
38 format = getFormat(mold
= [
0._TKG], ed
= SK_
"es", signed
= .true._LK)
41 call disp%show(
"ndim = getUnifRand(1, 7)")
45 call disp%show(
"std = getUnifRand(1, 10, ndim)")
48 call disp%show( std ,
format = format )
49 call disp%show(
"cor = getCovRand(1., ndim)")
52 call disp%show( cor ,
format = format )
53 call disp%show(
"cov = getFilled(0._TKG, ndim, ndim + 1)")
55 call disp%show(
"call setMatCopy(cov(1:ndim, 1:ndim), rdpack, cor, rdpack, lowDia)")
56 call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
58 call disp%show( cov ,
format = format )
59 call disp%show(
"call setCov(cov(1:ndim, 2:ndim+1), uppDia, cov(1:ndim, 1:ndim), lowDia, std) ! convert lower-triangle correlation matrix to upper-triangle covariance matrix.")
60 call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
62 call disp%show( cov ,
format = format )
64 call disp%show(
"cov = getFilled(0._TKG, ndim, ndim + 1)")
66 call disp%show(
"call setMatCopy(cov(1:ndim, 2:ndim+1), rdpack, cor, rdpack, uppDia)")
67 call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
69 call disp%show( cov ,
format = format )
70 call disp%show(
"call setCov(cov(1:ndim, 1:ndim), lowDia, cov(1:ndim, 2:ndim+1), uppDia, std) ! convert upper-triangle correlation matrix to lower-triangle covariance matrix.")
71 call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
73 call disp%show( cov ,
format = format )
79 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
80 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
81 call disp%show(
"!Compute the covariance matrix of a 2-D sample.")
82 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
83 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
88 real(TKG),
allocatable :: sample(:,:), samShifted(:,:), cov(:,:), mean(:)
89 integer(IK) :: ndim, nsam, dim
90 call disp%show(
"ndim = 2; nsam = 10; dim = 2")
91 ndim
= 2; nsam
= 10; dim
= 2
92 call disp%show(
"sample = reshape(getUnifRand(1, 20, ndim * nsam), shape = [ndim, nsam], order = [2, 1])")
93 sample
= reshape(
getUnifRand(
1,
20, ndim
* nsam), shape
= [ndim, nsam], order
= [
2,
1])
96 call disp%show(
"mean = getMean(sample, dim)")
100 call disp%show(
"samShifted = getShifted(sample, dim, -mean)")
104 call disp%show(
"cov = getFilled(0., ndim, ndim)")
106 call disp%show(
"call setCov(cov, uppDia, mean, sample, dim)")
107 call setCov(cov, uppDia, mean, sample, dim)
110 call disp%show(
"cov = getFilled(0., ndim, ndim)")
112 call disp%show(
"call setCov(cov, uppDia, samShifted, dim) ! same result as above.")
113 call setCov(cov, uppDia, samShifted, dim)
117 call disp%show(
"cov = getFilled(0., ndim, ndim)")
119 call disp%show(
"call setCov(cov, lowDia, mean, sample, dim)")
120 call setCov(cov, lowDia, mean, sample, dim)
123 call disp%show(
"cov = getFilled(0., ndim, ndim)")
125 call disp%show(
"call setCov(cov, lowDia, samShifted, dim) ! same result as above.")
126 call setCov(cov, lowDia, samShifted, dim)
130 call disp%show(
"Compute the sample covariance along the first dimension.", deliml
= SK_
'''')
134 call disp%show(
"cov = getFilled(0., ndim, ndim)")
136 call disp%show(
"call setCov(cov, uppDia, mean, transpose(sample), dim)")
137 call setCov(cov, uppDia, mean,
transpose(sample), dim)
140 call disp%show(
"cov = getFilled(0., ndim, ndim)")
142 call disp%show(
"call setCov(cov, uppDia, transpose(samShifted), dim) ! same result as above.")
143 call setCov(cov, uppDia,
transpose(samShifted), dim)
147 call disp%show(
"cov = getFilled(0., ndim, ndim)")
149 call disp%show(
"call setCov(cov, lowDia, mean, transpose(sample), dim)")
150 call setCov(cov, lowDia, mean,
transpose(sample), dim)
153 call disp%show(
"cov = getFilled(0., ndim, ndim)")
155 call disp%show(
"call setCov(cov, lowDia, transpose(samShifted), dim) ! same result as above.")
156 call setCov(cov, lowDia,
transpose(samShifted), dim)
160 call disp%show(
"Compute the full sample covariance for a pair of time series.", deliml
= SK_
'''')
162 call disp%show(
"cov = getFilled(0., ndim, ndim)")
164 call disp%show(
"call setCov(cov, mean, sample(1,:), sample(2,:))")
165 call setCov(cov, mean, sample(
1,:), sample(
2,:))
168 call disp%show(
"cov = getFilled(0., ndim, ndim)")
170 call disp%show(
"call setCov(cov, samShifted(1,:), samShifted(2,:)) ! same result as above.")
171 call setCov(cov, samShifted(
1,:), samShifted(
2,:))
178 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
179 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
180 call disp%show(
"!Compute the biased covariance matrix of a weighted 2-D sample.")
181 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
182 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
188 integer(IK) :: iweisum
189 real(TKG),
allocatable :: rweight(:)
190 integer(IK),
allocatable :: iweight(:)
191 real(TKG),
allocatable :: sample(:,:), samShifted(:,:), cov(:,:), mean(:)
192 integer(IK) :: ndim, nsam, dim
193 call disp%show(
"ndim = 2; nsam = 10; dim = 2")
194 ndim
= 2; nsam
= 10; dim
= 2
195 call disp%show(
"sample = reshape(getUnifRand(1, 20, ndim * nsam), shape = [ndim, nsam], order = [2, 1])")
196 sample
= reshape(
getUnifRand(
1,
20, ndim
* nsam), shape
= [ndim, nsam], order
= [
2,
1])
199 call disp%show(
"call setResized(mean, ndim)")
201 call disp%show(
"iweight = getUnifRand(1, 10, nsam) ! integer-valued weights.")
205 call disp%show(
"call setMean(mean, sample, dim, iweight, iweisum)")
206 call setMean(mean, sample, dim, iweight, iweisum)
211 call disp%show(
"rweight = iweight ! or real-valued weights.")
215 call disp%show(
"call setMean(mean, sample, dim, rweight, rweisum)")
216 call setMean(mean, sample, dim, rweight, rweisum)
223 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
224 call disp%show(
"!Compute the covariance matrix integer weights.")
225 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
228 call disp%show(
"samShifted = getShifted(sample, dim, -mean)")
232 call disp%show(
"cov = getFilled(0., ndim, ndim)")
234 call disp%show(
"call setCov(cov, uppDia, mean, sample, dim, iweight, iweisum)")
235 call setCov(cov, uppDia, mean, sample, dim, iweight, iweisum)
238 call disp%show(
"cov = getFilled(0., ndim, ndim)")
240 call disp%show(
"call setCov(cov, uppDia, samShifted, dim, iweight, sum(iweight)) ! same result as above.")
241 call setCov(cov, uppDia, samShifted, dim, iweight,
sum(iweight))
245 call disp%show(
"cov = getFilled(0., ndim, ndim)")
247 call disp%show(
"call setCov(cov, lowDia, mean, sample, dim, iweight, iweisum)")
248 call setCov(cov, lowDia, mean, sample, dim, iweight, iweisum)
251 call disp%show(
"cov = getFilled(0., ndim, ndim)")
253 call disp%show(
"call setCov(cov, lowDia, samShifted, dim, iweight, sum(iweight)) ! same result as above.")
254 call setCov(cov, lowDia, samShifted, dim, iweight,
sum(iweight))
258 call disp%show(
"Compute the sample covariance along the first dimension.", deliml
= SK_
'''')
262 call disp%show(
"cov = getFilled(0., ndim, ndim)")
264 call disp%show(
"call setCov(cov, uppDia, mean, transpose(sample), dim, iweight, iweisum)")
265 call setCov(cov, uppDia, mean,
transpose(sample), dim, iweight, iweisum)
268 call disp%show(
"cov = getFilled(0., ndim, ndim)")
270 call disp%show(
"call setCov(cov, uppDia, transpose(samShifted), dim, iweight, sum(iweight)) ! same result as above.")
271 call setCov(cov, uppDia,
transpose(samShifted), dim, iweight,
sum(iweight))
275 call disp%show(
"cov = getFilled(0., ndim, ndim)")
277 call disp%show(
"call setCov(cov, lowDia, mean, transpose(sample), dim, iweight, iweisum)")
278 call setCov(cov, lowDia, mean,
transpose(sample), dim, iweight, iweisum)
281 call disp%show(
"cov = getFilled(0., ndim, ndim)")
283 call disp%show(
"call setCov(cov, lowDia, transpose(samShifted), dim, iweight, sum(iweight)) ! same result as above.")
284 call setCov(cov, lowDia,
transpose(samShifted), dim, iweight,
sum(iweight))
288 call disp%show(
"Compute the full sample covariance for a pair of time series.", deliml
= SK_
'''')
290 call disp%show(
"cov = getFilled(0., ndim, ndim)")
292 call disp%show(
"call setCov(cov, mean, sample(1,:), sample(2,:), iweight, iweisum)")
293 call setCov(cov, mean, sample(
1,:), sample(
2,:), iweight, iweisum)
296 call disp%show(
"cov = getFilled(0., ndim, ndim)")
298 call disp%show(
"call setCov(cov, samShifted(1,:), samShifted(2,:), iweight, sum(iweight)) ! same result as above.")
299 call setCov(cov, samShifted(
1,:), samShifted(
2,:), iweight,
sum(iweight))
305 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
306 call disp%show(
"!Compute the covariance matrix real weights.")
307 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
312 call disp%show(
"samShifted = getShifted(sample, dim, -mean)")
316 call disp%show(
"cov = getFilled(0., ndim, ndim)")
318 call disp%show(
"call setCov(cov, uppDia, mean, sample, dim, rweight, rweisum)")
319 call setCov(cov, uppDia, mean, sample, dim, rweight, rweisum)
322 call disp%show(
"cov = getFilled(0., ndim, ndim)")
324 call disp%show(
"call setCov(cov, uppDia, samShifted, dim, rweight, sum(rweight)) ! same result as above.")
325 call setCov(cov, uppDia, samShifted, dim, rweight,
sum(rweight))
329 call disp%show(
"cov = getFilled(0., ndim, ndim)")
331 call disp%show(
"call setCov(cov, lowDia, mean, sample, dim, rweight, rweisum)")
332 call setCov(cov, lowDia, mean, sample, dim, rweight, rweisum)
335 call disp%show(
"cov = getFilled(0., ndim, ndim)")
337 call disp%show(
"call setCov(cov, lowDia, samShifted, dim, rweight, sum(rweight)) ! same result as above.")
338 call setCov(cov, lowDia, samShifted, dim, rweight,
sum(rweight))
342 call disp%show(
"Compute the sample covariance along the first dimension.", deliml
= SK_
'''')
346 call disp%show(
"cov = getFilled(0., ndim, ndim)")
348 call disp%show(
"call setCov(cov, uppDia, mean, transpose(sample), dim, rweight, rweisum)")
349 call setCov(cov, uppDia, mean,
transpose(sample), dim, rweight, rweisum)
352 call disp%show(
"cov = getFilled(0., ndim, ndim)")
354 call disp%show(
"call setCov(cov, uppDia, transpose(samShifted), dim, rweight, sum(rweight)) ! same result as above.")
355 call setCov(cov, uppDia,
transpose(samShifted), dim, rweight,
sum(rweight))
359 call disp%show(
"cov = getFilled(0., ndim, ndim)")
361 call disp%show(
"call setCov(cov, lowDia, mean, transpose(sample), dim, rweight, rweisum)")
362 call setCov(cov, lowDia, mean,
transpose(sample), dim, rweight, rweisum)
365 call disp%show(
"cov = getFilled(0., ndim, ndim)")
367 call disp%show(
"call setCov(cov, lowDia, transpose(samShifted), dim, rweight, sum(rweight)) ! same result as above.")
368 call setCov(cov, lowDia,
transpose(samShifted), dim, rweight,
sum(rweight))
372 call disp%show(
"Compute the full sample covariance for a pair of time series.", deliml
= SK_
'''')
374 call disp%show(
"cov = getFilled(0., ndim, ndim)")
376 call disp%show(
"call setCov(cov, mean, sample(1,:), sample(2,:), rweight, rweisum)")
377 call setCov(cov, mean, sample(
1,:), sample(
2,:), rweight, rweisum)
380 call disp%show(
"cov = getFilled(0., ndim, ndim)")
382 call disp%show(
"call setCov(cov, samShifted(1,:), samShifted(2,:), rweight, sum(rweight)) ! same result as above.")
383 call setCov(cov, samShifted(
1,:), samShifted(
2,:), rweight,
sum(rweight))
Generate and return an array of the specified rank and shape of arbitrary intrinsic type and kind wit...
Allocate or resize (shrink or expand) an input allocatable scalar string or array of rank 1....
Generate count evenly spaced points over the interval [x1, x2] if x1 < x2, or [x2,...
Generate and return a random positive-definite (correlation or covariance) matrix using the Gram meth...
Generate and return a scalar or a contiguous array of rank 1 of length s1 of randomly uniformly distr...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
Copy a desired subset of the input source matrix of arbitrary shape (:) or (:,:) to the target subset...
Generate and return the (weighted) mean of an input sample of nsam observations with ndim = 1 or 2 at...
Return the (weighted) mean of a pair of time series or of an input sample of nsam observations with n...
Generate a sample of shape (nsam), or (ndim, nsam) or (nsam, ndim) that is shifted by the specified i...
This module contains procedures and generic interfaces for convenient allocation and filling of array...
This module contains procedures and generic interfaces for resizing allocatable arrays of various typ...
This module contains procedures and generic interfaces for generating arrays with linear or logarithm...
This module contains classes and procedures for generating random matrices distributed on the space o...
This module contains classes and procedures for computing various statistical quantities related to t...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
integer, parameter RKS
The single-precision real kind in Fortran mode. On most platforms, this is an 32-bit real kind.
This module contains procedures and generic interfaces relevant to copying (diagonal or upper/lower t...
This module contains classes and procedures for computing the first moment (i.e., the statistical mea...
This module contains classes and procedures for shifting univariate or multivariate samples by arbitr...
Generate and return an object of type display_type.
14+4.000000E+00,
+2.000000E+00
17+1.000000E+00,
-7.630376E-01
18-7.630376E-01,
+1.000000E+00
20call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
22+1.000000E+00,
+0.000000E+00,
+0.000000E+00
23-7.630376E-01,
+1.000000E+00,
+0.000000E+00
24call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
26+1.000000E+00,
+1.600000E+01,
-6.104300E+00
27-7.630376E-01,
+1.000000E+00,
+4.000000E+00
30call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
32+0.000000E+00,
+1.000000E+00,
-7.630376E-01
33+0.000000E+00,
+0.000000E+00,
+1.000000E+00
34call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
36+1.600000E+01,
+1.000000E+00,
-7.630376E-01
37-6.104300E+00,
+4.000000E+00,
+1.000000E+00
45+5.000000E+00,
+5.000000E+00,
+1.000000E+00,
+1.000000E+01
48+1.000000E+00,
+2.005533E-01,
-1.883450E-01,
+1.922954E-01
49+2.005533E-01,
+1.000000E+00,
-6.863858E-01,
+2.038796E-02
50-1.883450E-01,
-6.863858E-01,
+1.000000E+00,
-7.081194E-01
51+1.922954E-01,
+2.038796E-02,
-7.081194E-01,
+1.000000E+00
53call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
55+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
56+2.005533E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
57-1.883450E-01,
-6.863858E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
58+1.922954E-01,
+2.038796E-02,
-7.081194E-01,
+1.000000E+00,
+0.000000E+00
59call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
61+1.000000E+00,
+2.500000E+01,
+5.013832E+00,
-9.417253E-01,
+9.614773E+00
62+2.005533E-01,
+1.000000E+00,
+2.500000E+01,
-3.431929E+00,
+1.019398E+00
63-1.883450E-01,
-6.863858E-01,
+1.000000E+00,
+1.000000E+00,
-7.081194E+00
64+1.922954E-01,
+2.038796E-02,
-7.081194E-01,
+1.000000E+00,
+1.000000E+02
67call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
69+0.000000E+00,
+1.000000E+00,
+2.005533E-01,
-1.883450E-01,
+1.922954E-01
70+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-6.863858E-01,
+2.038796E-02
71+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-7.081194E-01
72+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
73call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
75+2.500000E+01,
+1.000000E+00,
+2.005533E-01,
-1.883450E-01,
+1.922954E-01
76+5.013832E+00,
+2.500000E+01,
+1.000000E+00,
-6.863858E-01,
+2.038796E-02
77-9.417253E-01,
-3.431929E+00,
+1.000000E+00,
+1.000000E+00,
-7.081194E-01
78+9.614773E+00,
+1.019398E+00,
-7.081194E+00,
+1.000000E+02,
+1.000000E+00
86+5.000000E+00,
+2.000000E+00,
+4.000000E+00
89+1.000000E+00,
+5.682755E-02,
-2.933485E-01
90+5.682755E-02,
+1.000000E+00,
+6.507880E-01
91-2.933485E-01,
+6.507880E-01,
+1.000000E+00
93call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
95+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
96+5.682755E-02,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
97-2.933485E-01,
+6.507880E-01,
+1.000000E+00,
+0.000000E+00
98call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
100+1.000000E+00,
+2.500000E+01,
+5.682755E-01,
-5.866969E+00
101+5.682755E-02,
+1.000000E+00,
+4.000000E+00,
+5.206304E+00
102-2.933485E-01,
+6.507880E-01,
+1.000000E+00,
+1.600000E+01
105call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
107+0.000000E+00,
+1.000000E+00,
+5.682755E-02,
-2.933485E-01
108+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+6.507880E-01
109+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
110call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
112+2.500000E+01,
+1.000000E+00,
+5.682755E-02,
-2.933485E-01
113+5.682755E-01,
+4.000000E+00,
+1.000000E+00,
+6.507880E-01
114-5.866969E+00,
+5.206304E+00,
+1.600000E+01,
+1.000000E+00
122+7.000000E+00,
+8.000000E+00
125+1.000000E+00,
+6.787896E-01
126+6.787896E-01,
+1.000000E+00
128call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
130+1.000000E+00,
+0.000000E+00,
+0.000000E+00
131+6.787896E-01,
+1.000000E+00,
+0.000000E+00
132call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
134+1.000000E+00,
+4.900000E+01,
+3.801222E+01
135+6.787896E-01,
+1.000000E+00,
+6.400000E+01
138call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
140+0.000000E+00,
+1.000000E+00,
+6.787896E-01
141+0.000000E+00,
+0.000000E+00,
+1.000000E+00
142call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
144+4.900000E+01,
+1.000000E+00,
+6.787896E-01
145+3.801222E+01,
+6.400000E+01,
+1.000000E+00
153+1.000000E+01,
+7.000000E+00,
+1.000000E+01,
+9.000000E+00,
+1.000000E+00
156+1.000000E+00,
+1.674978E-01,
+3.232417E-01,
-2.522079E-02,
+4.693460E-01
157+1.674978E-01,
+1.000000E+00,
-7.061831E-01,
-5.579556E-01,
+1.891168E-01
158+3.232417E-01,
-7.061831E-01,
+1.000000E+00,
+6.764400E-01,
-2.016856E-01
159-2.522079E-02,
-5.579556E-01,
+6.764400E-01,
+1.000000E+00,
-7.950259E-01
160+4.693460E-01,
+1.891168E-01,
-2.016856E-01,
-7.950259E-01,
+1.000000E+00
162call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
164+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
165+1.674978E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
166+3.232417E-01,
-7.061831E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
167-2.522079E-02,
-5.579556E-01,
+6.764400E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
168+4.693460E-01,
+1.891168E-01,
-2.016856E-01,
-7.950259E-01,
+1.000000E+00,
+0.000000E+00
169call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
171+1.000000E+00,
+1.000000E+02,
+1.172485E+01,
+3.232417E+01,
-2.269871E+00,
+4.693460E+00
172+1.674978E-01,
+1.000000E+00,
+4.900000E+01,
-4.943282E+01,
-3.515120E+01,
+1.323818E+00
173+3.232417E-01,
-7.061831E-01,
+1.000000E+00,
+1.000000E+02,
+6.087960E+01,
-2.016856E+00
174-2.522079E-02,
-5.579556E-01,
+6.764400E-01,
+1.000000E+00,
+8.100000E+01,
-7.155233E+00
175+4.693460E-01,
+1.891168E-01,
-2.016856E-01,
-7.950259E-01,
+1.000000E+00,
+1.000000E+00
178call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
180+0.000000E+00,
+1.000000E+00,
+1.674978E-01,
+3.232417E-01,
-2.522079E-02,
+4.693460E-01
181+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-7.061831E-01,
-5.579556E-01,
+1.891168E-01
182+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+6.764400E-01,
-2.016856E-01
183+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-7.950259E-01
184+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
185call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
187+1.000000E+02,
+1.000000E+00,
+1.674978E-01,
+3.232417E-01,
-2.522079E-02,
+4.693460E-01
188+1.172485E+01,
+4.900000E+01,
+1.000000E+00,
-7.061831E-01,
-5.579556E-01,
+1.891168E-01
189+3.232417E+01,
-4.943282E+01,
+1.000000E+02,
+1.000000E+00,
+6.764400E-01,
-2.016856E-01
190-2.269871E+00,
-3.515120E+01,
+6.087960E+01,
+8.100000E+01,
+1.000000E+00,
-7.950259E-01
191+4.693460E+00,
+1.323818E+00,
-2.016856E+00,
-7.155233E+00,
+1.000000E+00,
+1.000000E+00
199+1.000000E+00,
+2.000000E+00,
+4.000000E+00,
+7.000000E+00,
+1.000000E+01,
+5.000000E+00,
+6.000000E+00
202+1.000000E+00,
-8.015262E-01,
+3.629454E-01,
-2.950583E-01,
+5.747038E-01,
-5.639211E-01,
-2.825486E-02
203-8.015262E-01,
+1.000000E+00,
-3.324285E-01,
+4.862410E-01,
-7.673784E-01,
+4.132438E-01,
-9.036963E-02
204+3.629454E-01,
-3.324285E-01,
+1.000000E+00,
-2.069316E-01,
+7.475872E-01,
-7.230764E-01,
+5.369414E-01
205-2.950583E-01,
+4.862410E-01,
-2.069316E-01,
+1.000000E+00,
-1.723250E-01,
+5.715978E-01,
-2.411085E-01
206+5.747038E-01,
-7.673784E-01,
+7.475872E-01,
-1.723250E-01,
+1.000000E+00,
-4.669105E-01,
+2.909592E-01
207-5.639211E-01,
+4.132438E-01,
-7.230764E-01,
+5.715978E-01,
-4.669105E-01,
+1.000000E+00,
-1.626980E-01
208-2.825486E-02,
-9.036963E-02,
+5.369414E-01,
-2.411085E-01,
+2.909592E-01,
-1.626980E-01,
+1.000000E+00
210call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
212+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
213-8.015262E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
214+3.629454E-01,
-3.324285E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
215-2.950583E-01,
+4.862410E-01,
-2.069316E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
216+5.747038E-01,
-7.673784E-01,
+7.475872E-01,
-1.723250E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
217-5.639211E-01,
+4.132438E-01,
-7.230764E-01,
+5.715978E-01,
-4.669105E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
218-2.825486E-02,
-9.036963E-02,
+5.369414E-01,
-2.411085E-01,
+2.909592E-01,
-1.626980E-01,
+1.000000E+00,
+0.000000E+00
219call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
221+1.000000E+00,
+1.000000E+00,
-1.603052E+00,
+1.451782E+00,
-2.065408E+00,
+5.747038E+00,
-2.819605E+00,
-1.695291E-01
222-8.015262E-01,
+1.000000E+00,
+4.000000E+00,
-2.659428E+00,
+6.807374E+00,
-1.534757E+01,
+4.132438E+00,
-1.084435E+00
223+3.629454E-01,
-3.324285E-01,
+1.000000E+00,
+1.600000E+01,
-5.794084E+00,
+2.990349E+01,
-1.446153E+01,
+1.288659E+01
224-2.950583E-01,
+4.862410E-01,
-2.069316E-01,
+1.000000E+00,
+4.900000E+01,
-1.206275E+01,
+2.000592E+01,
-1.012656E+01
225+5.747038E-01,
-7.673784E-01,
+7.475872E-01,
-1.723250E-01,
+1.000000E+00,
+1.000000E+02,
-2.334552E+01,
+1.745755E+01
226-5.639211E-01,
+4.132438E-01,
-7.230764E-01,
+5.715978E-01,
-4.669105E-01,
+1.000000E+00,
+2.500000E+01,
-4.880940E+00
227-2.825486E-02,
-9.036963E-02,
+5.369414E-01,
-2.411085E-01,
+2.909592E-01,
-1.626980E-01,
+1.000000E+00,
+3.600000E+01
230call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
232+0.000000E+00,
+1.000000E+00,
-8.015262E-01,
+3.629454E-01,
-2.950583E-01,
+5.747038E-01,
-5.639211E-01,
-2.825486E-02
233+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-3.324285E-01,
+4.862410E-01,
-7.673784E-01,
+4.132438E-01,
-9.036963E-02
234+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-2.069316E-01,
+7.475872E-01,
-7.230764E-01,
+5.369414E-01
235+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-1.723250E-01,
+5.715978E-01,
-2.411085E-01
236+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-4.669105E-01,
+2.909592E-01
237+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-1.626980E-01
238+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
239call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
241+1.000000E+00,
+1.000000E+00,
-8.015262E-01,
+3.629454E-01,
-2.950583E-01,
+5.747038E-01,
-5.639211E-01,
-2.825486E-02
242-1.603052E+00,
+4.000000E+00,
+1.000000E+00,
-3.324285E-01,
+4.862410E-01,
-7.673784E-01,
+4.132438E-01,
-9.036963E-02
243+1.451782E+00,
-2.659428E+00,
+1.600000E+01,
+1.000000E+00,
-2.069316E-01,
+7.475872E-01,
-7.230764E-01,
+5.369414E-01
244-2.065408E+00,
+6.807374E+00,
-5.794084E+00,
+4.900000E+01,
+1.000000E+00,
-1.723250E-01,
+5.715978E-01,
-2.411085E-01
245+5.747038E+00,
-1.534757E+01,
+2.990349E+01,
-1.206275E+01,
+1.000000E+02,
+1.000000E+00,
-4.669105E-01,
+2.909592E-01
246-2.819605E+00,
+4.132438E+00,
-1.446153E+01,
+2.000592E+01,
-2.334552E+01,
+2.500000E+01,
+1.000000E+00,
-1.626980E-01
247-1.695291E-01,
-1.084435E+00,
+1.288659E+01,
-1.012656E+01,
+1.745755E+01,
-4.880940E+00,
+3.600000E+01,
+1.000000E+00
255+9.000000E+00,
+5.000000E+00,
+6.000000E+00,
+3.000000E+00,
+5.000000E+00,
+6.000000E+00
258+1.000000E+00,
-8.654518E-01,
-7.282782E-01,
+9.898201E-02,
+1.921818E-01,
+5.303325E-01
259-8.654518E-01,
+1.000000E+00,
+6.867865E-01,
-5.515494E-01,
-1.489851E-02,
-2.569512E-01
260-7.282782E-01,
+6.867865E-01,
+1.000000E+00,
-3.788385E-01,
-1.680465E-01,
-6.524359E-01
261+9.898201E-02,
-5.515494E-01,
-3.788385E-01,
+1.000000E+00,
-3.473597E-01,
-2.055989E-01
262+1.921818E-01,
-1.489851E-02,
-1.680465E-01,
-3.473597E-01,
+1.000000E+00,
+7.154284E-01
263+5.303325E-01,
-2.569512E-01,
-6.524359E-01,
-2.055989E-01,
+7.154284E-01,
+1.000000E+00
265call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
267+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
268-8.654518E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
269-7.282782E-01,
+6.867865E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
270+9.898201E-02,
-5.515494E-01,
-3.788385E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
271+1.921818E-01,
-1.489851E-02,
-1.680465E-01,
-3.473597E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
272+5.303325E-01,
-2.569512E-01,
-6.524359E-01,
-2.055989E-01,
+7.154284E-01,
+1.000000E+00,
+0.000000E+00
273call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
275+1.000000E+00,
+8.100000E+01,
-3.894533E+01,
-3.932702E+01,
+2.672514E+00,
+8.648179E+00,
+2.863795E+01
276-8.654518E-01,
+1.000000E+00,
+2.500000E+01,
+2.060359E+01,
-8.273241E+00,
-3.724627E-01,
-7.708535E+00
277-7.282782E-01,
+6.867865E-01,
+1.000000E+00,
+3.600000E+01,
-6.819093E+00,
-5.041395E+00,
-2.348769E+01
278+9.898201E-02,
-5.515494E-01,
-3.788385E-01,
+1.000000E+00,
+9.000000E+00,
-5.210396E+00,
-3.700780E+00
279+1.921818E-01,
-1.489851E-02,
-1.680465E-01,
-3.473597E-01,
+1.000000E+00,
+2.500000E+01,
+2.146285E+01
280+5.303325E-01,
-2.569512E-01,
-6.524359E-01,
-2.055989E-01,
+7.154284E-01,
+1.000000E+00,
+3.600000E+01
283call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
285+0.000000E+00,
+1.000000E+00,
-8.654518E-01,
-7.282782E-01,
+9.898201E-02,
+1.921818E-01,
+5.303325E-01
286+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+6.867865E-01,
-5.515494E-01,
-1.489851E-02,
-2.569512E-01
287+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-3.788385E-01,
-1.680465E-01,
-6.524359E-01
288+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-3.473597E-01,
-2.055989E-01
289+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+7.154284E-01
290+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
291call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
293+8.100000E+01,
+1.000000E+00,
-8.654518E-01,
-7.282782E-01,
+9.898201E-02,
+1.921818E-01,
+5.303325E-01
294-3.894533E+01,
+2.500000E+01,
+1.000000E+00,
+6.867865E-01,
-5.515494E-01,
-1.489851E-02,
-2.569512E-01
295-3.932702E+01,
+2.060359E+01,
+3.600000E+01,
+1.000000E+00,
-3.788385E-01,
-1.680465E-01,
-6.524359E-01
296+2.672514E+00,
-8.273241E+00,
-6.819093E+00,
+9.000000E+00,
+1.000000E+00,
-3.473597E-01,
-2.055989E-01
297+8.648179E+00,
-3.724627E-01,
-5.041395E+00,
-5.210396E+00,
+2.500000E+01,
+1.000000E+00,
+7.154284E-01
298+2.863795E+01,
-7.708535E+00,
-2.348769E+01,
-3.700780E+00,
+2.146285E+01,
+3.600000E+01,
+1.000000E+00
306+9.000000E+00,
+2.000000E+00,
+1.000000E+00,
+2.000000E+00
309+1.000000E+00,
-9.067490E-01,
+2.879211E-01,
-2.249667E-01
310-9.067490E-01,
+1.000000E+00,
-6.610098E-01,
+8.290101E-02
311+2.879211E-01,
-6.610098E-01,
+1.000000E+00,
+3.001972E-01
312-2.249667E-01,
+8.290101E-02,
+3.001972E-01,
+1.000000E+00
314call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
316+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
317-9.067490E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
318+2.879211E-01,
-6.610098E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
319-2.249667E-01,
+8.290101E-02,
+3.001972E-01,
+1.000000E+00,
+0.000000E+00
320call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
322+1.000000E+00,
+8.100000E+01,
-1.632148E+01,
+2.591290E+00,
-4.049401E+00
323-9.067490E-01,
+1.000000E+00,
+4.000000E+00,
-1.322020E+00,
+3.316040E-01
324+2.879211E-01,
-6.610098E-01,
+1.000000E+00,
+1.000000E+00,
+6.003944E-01
325-2.249667E-01,
+8.290101E-02,
+3.001972E-01,
+1.000000E+00,
+4.000000E+00
328call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
330+0.000000E+00,
+1.000000E+00,
-9.067490E-01,
+2.879211E-01,
-2.249667E-01
331+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-6.610098E-01,
+8.290101E-02
332+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+3.001972E-01
333+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
334call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
336+8.100000E+01,
+1.000000E+00,
-9.067490E-01,
+2.879211E-01,
-2.249667E-01
337-1.632148E+01,
+4.000000E+00,
+1.000000E+00,
-6.610098E-01,
+8.290101E-02
338+2.591290E+00,
-1.322020E+00,
+1.000000E+00,
+1.000000E+00,
+3.001972E-01
339-4.049401E+00,
+3.316040E-01,
+6.003944E-01,
+4.000000E+00,
+1.000000E+00
347+1.000000E+00,
+7.000000E+00,
+4.000000E+00,
+6.000000E+00,
+9.000000E+00,
+4.000000E+00,
+8.000000E+00
350+1.000000E+00,
+8.171124E-02,
+1.833007E-01,
-5.452252E-01,
+4.400409E-01,
+4.064409E-01,
+3.405425E-02
351+8.171124E-02,
+1.000000E+00,
+3.496451E-01,
+3.681374E-01,
-1.257106E-01,
+6.347072E-01,
-2.514751E-01
352+1.833007E-01,
+3.496451E-01,
+1.000000E+00,
+4.623571E-01,
+4.696944E-01,
+6.954970E-01,
-1.163978E-01
353-5.452252E-01,
+3.681374E-01,
+4.623571E-01,
+1.000000E+00,
-3.147775E-01,
+3.116178E-01,
-4.606161E-01
354+4.400409E-01,
-1.257106E-01,
+4.696944E-01,
-3.147775E-01,
+1.000000E+00,
+3.610232E-01,
-2.773589E-02
355+4.064409E-01,
+6.347072E-01,
+6.954970E-01,
+3.116178E-01,
+3.610232E-01,
+1.000000E+00,
-1.573637E-01
356+3.405425E-02,
-2.514751E-01,
-1.163978E-01,
-4.606161E-01,
-2.773589E-02,
-1.573637E-01,
+1.000000E+00
358call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
360+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
361+8.171124E-02,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
362+1.833007E-01,
+3.496451E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
363-5.452252E-01,
+3.681374E-01,
+4.623571E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
364+4.400409E-01,
-1.257106E-01,
+4.696944E-01,
-3.147775E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
365+4.064409E-01,
+6.347072E-01,
+6.954970E-01,
+3.116178E-01,
+3.610232E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
366+3.405425E-02,
-2.514751E-01,
-1.163978E-01,
-4.606161E-01,
-2.773589E-02,
-1.573637E-01,
+1.000000E+00,
+0.000000E+00
367call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
369+1.000000E+00,
+1.000000E+00,
+5.719787E-01,
+7.332030E-01,
-3.271351E+00,
+3.960368E+00,
+1.625764E+00,
+2.724340E-01
370+8.171124E-02,
+1.000000E+00,
+4.900000E+01,
+9.790064E+00,
+1.546177E+01,
-7.919768E+00,
+1.777180E+01,
-1.408260E+01
371+1.833007E-01,
+3.496451E-01,
+1.000000E+00,
+1.600000E+01,
+1.109657E+01,
+1.690900E+01,
+1.112795E+01,
-3.724731E+00
372-5.452252E-01,
+3.681374E-01,
+4.623571E-01,
+1.000000E+00,
+3.600000E+01,
-1.699798E+01,
+7.478827E+00,
-2.210957E+01
373+4.400409E-01,
-1.257106E-01,
+4.696944E-01,
-3.147775E-01,
+1.000000E+00,
+8.100000E+01,
+1.299683E+01,
-1.996984E+00
374+4.064409E-01,
+6.347072E-01,
+6.954970E-01,
+3.116178E-01,
+3.610232E-01,
+1.000000E+00,
+1.600000E+01,
-5.035639E+00
375+3.405425E-02,
-2.514751E-01,
-1.163978E-01,
-4.606161E-01,
-2.773589E-02,
-1.573637E-01,
+1.000000E+00,
+6.400000E+01
378call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
380+0.000000E+00,
+1.000000E+00,
+8.171124E-02,
+1.833007E-01,
-5.452252E-01,
+4.400409E-01,
+4.064409E-01,
+3.405425E-02
381+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+3.496451E-01,
+3.681374E-01,
-1.257106E-01,
+6.347072E-01,
-2.514751E-01
382+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+4.623571E-01,
+4.696944E-01,
+6.954970E-01,
-1.163978E-01
383+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-3.147775E-01,
+3.116178E-01,
-4.606161E-01
384+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+3.610232E-01,
-2.773589E-02
385+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-1.573637E-01
386+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
387call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
389+1.000000E+00,
+1.000000E+00,
+8.171124E-02,
+1.833007E-01,
-5.452252E-01,
+4.400409E-01,
+4.064409E-01,
+3.405425E-02
390+5.719787E-01,
+4.900000E+01,
+1.000000E+00,
+3.496451E-01,
+3.681374E-01,
-1.257106E-01,
+6.347072E-01,
-2.514751E-01
391+7.332030E-01,
+9.790064E+00,
+1.600000E+01,
+1.000000E+00,
+4.623571E-01,
+4.696944E-01,
+6.954970E-01,
-1.163978E-01
392-3.271351E+00,
+1.546177E+01,
+1.109657E+01,
+3.600000E+01,
+1.000000E+00,
-3.147775E-01,
+3.116178E-01,
-4.606161E-01
393+3.960368E+00,
-7.919768E+00,
+1.690900E+01,
-1.699798E+01,
+8.100000E+01,
+1.000000E+00,
+3.610232E-01,
-2.773589E-02
394+1.625764E+00,
+1.777180E+01,
+1.112795E+01,
+7.478827E+00,
+1.299683E+01,
+1.600000E+01,
+1.000000E+00,
-1.573637E-01
395+2.724340E-01,
-1.408260E+01,
-3.724731E+00,
-2.210957E+01,
-1.996984E+00,
-5.035639E+00,
+6.400000E+01,
+1.000000E+00
403+3.000000E+00,
+1.000000E+01,
+9.000000E+00,
+8.000000E+00,
+4.000000E+00,
+4.000000E+00,
+4.000000E+00
406+1.000000E+00,
+2.009096E-01,
-6.263593E-01,
+3.890767E-01,
+3.159687E-01,
+3.334182E-02,
-5.066900E-01
407+2.009096E-01,
+1.000000E+00,
-8.316587E-01,
-6.206609E-01,
-4.078837E-01,
+4.053683E-01,
-7.023707E-01
408-6.263593E-01,
-8.316587E-01,
+1.000000E+00,
+3.916197E-01,
+2.127269E-01,
-1.632651E-01,
+7.485247E-01
409+3.890767E-01,
-6.206609E-01,
+3.916197E-01,
+1.000000E+00,
+7.647105E-01,
-1.790522E-01,
+3.488088E-01
410+3.159687E-01,
-4.078837E-01,
+2.127269E-01,
+7.647105E-01,
+1.000000E+00,
-3.859851E-01,
+3.717218E-01
411+3.334182E-02,
+4.053683E-01,
-1.632651E-01,
-1.790522E-01,
-3.859851E-01,
+1.000000E+00,
-6.094661E-01
412-5.066900E-01,
-7.023707E-01,
+7.485247E-01,
+3.488088E-01,
+3.717218E-01,
-6.094661E-01,
+1.000000E+00
414call setMatCopy(cov(
1:ndim,
1:ndim), rdpack, cor, rdpack, lowDia)
416+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
417+2.009096E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
418-6.263593E-01,
-8.316587E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
419+3.890767E-01,
-6.206609E-01,
+3.916197E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
420+3.159687E-01,
-4.078837E-01,
+2.127269E-01,
+7.647105E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00
421+3.334182E-02,
+4.053683E-01,
-1.632651E-01,
-1.790522E-01,
-3.859851E-01,
+1.000000E+00,
+0.000000E+00,
+0.000000E+00
422-5.066900E-01,
-7.023707E-01,
+7.485247E-01,
+3.488088E-01,
+3.717218E-01,
-6.094661E-01,
+1.000000E+00,
+0.000000E+00
423call setCov(cov(
1:ndim,
2:ndim
+1), uppDia, cov(
1:ndim,
1:ndim), lowDia, std)
425+1.000000E+00,
+9.000000E+00,
+6.027289E+00,
-1.691170E+01,
+9.337840E+00,
+3.791625E+00,
+4.001018E-01,
-6.080279E+00
426+2.009096E-01,
+1.000000E+00,
+1.000000E+02,
-7.484928E+01,
-4.965287E+01,
-1.631535E+01,
+1.621473E+01,
-2.809483E+01
427-6.263593E-01,
-8.316587E-01,
+1.000000E+00,
+8.100000E+01,
+2.819662E+01,
+7.658167E+00,
-5.877543E+00,
+2.694689E+01
428+3.890767E-01,
-6.206609E-01,
+3.916197E-01,
+1.000000E+00,
+6.400000E+01,
+2.447074E+01,
-5.729670E+00,
+1.116188E+01
429+3.159687E-01,
-4.078837E-01,
+2.127269E-01,
+7.647105E-01,
+1.000000E+00,
+1.600000E+01,
-6.175761E+00,
+5.947549E+00
430+3.334182E-02,
+4.053683E-01,
-1.632651E-01,
-1.790522E-01,
-3.859851E-01,
+1.000000E+00,
+1.600000E+01,
-9.751458E+00
431-5.066900E-01,
-7.023707E-01,
+7.485247E-01,
+3.488088E-01,
+3.717218E-01,
-6.094661E-01,
+1.000000E+00,
+1.600000E+01
434call setMatCopy(cov(
1:ndim,
2:ndim
+1), rdpack, cor, rdpack, uppDia)
436+0.000000E+00,
+1.000000E+00,
+2.009096E-01,
-6.263593E-01,
+3.890767E-01,
+3.159687E-01,
+3.334182E-02,
-5.066900E-01
437+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-8.316587E-01,
-6.206609E-01,
-4.078837E-01,
+4.053683E-01,
-7.023707E-01
438+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+3.916197E-01,
+2.127269E-01,
-1.632651E-01,
+7.485247E-01
439+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
+7.647105E-01,
-1.790522E-01,
+3.488088E-01
440+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-3.859851E-01,
+3.717218E-01
441+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00,
-6.094661E-01
442+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+0.000000E+00,
+1.000000E+00
443call setCov(cov(
1:ndim,
1:ndim), lowDia, cov(
1:ndim,
2:ndim
+1), uppDia, std)
445+9.000000E+00,
+1.000000E+00,
+2.009096E-01,
-6.263593E-01,
+3.890767E-01,
+3.159687E-01,
+3.334182E-02,
-5.066900E-01
446+6.027289E+00,
+1.000000E+02,
+1.000000E+00,
-8.316587E-01,
-6.206609E-01,
-4.078837E-01,
+4.053683E-01,
-7.023707E-01
447-1.691170E+01,
-7.484928E+01,
+8.100000E+01,
+1.000000E+00,
+3.916197E-01,
+2.127269E-01,
-1.632651E-01,
+7.485247E-01
448+9.337840E+00,
-4.965287E+01,
+2.819662E+01,
+6.400000E+01,
+1.000000E+00,
+7.647105E-01,
-1.790522E-01,
+3.488088E-01
449+3.791625E+00,
-1.631535E+01,
+7.658167E+00,
+2.447074E+01,
+1.600000E+01,
+1.000000E+00,
-3.859851E-01,
+3.717218E-01
450+4.001018E-01,
+1.621473E+01,
-5.877543E+00,
-5.729670E+00,
-6.175761E+00,
+1.600000E+01,
+1.000000E+00,
-6.094661E-01
451-6.080279E+00,
-2.809483E+01,
+2.694689E+01,
+1.116188E+01,
+5.947549E+00,
-9.751458E+00,
+1.600000E+01,
+1.000000E+00
460ndim
= 2; nsam
= 10; dim
= 2
461sample
= reshape(
getUnifRand(
1,
20, ndim
* nsam), shape
= [ndim, nsam], order
= [
2,
1])
463+20.0000000,
+7.00000000,
+15.0000000,
+9.00000000,
+10.0000000,
+7.00000000,
+5.00000000,
+7.00000000,
+15.0000000,
+20.0000000
464+16.0000000,
+5.00000000,
+10.0000000,
+16.0000000,
+6.00000000,
+13.0000000,
+18.0000000,
+11.0000000,
+11.0000000,
+3.00000000
467+11.5000000,
+10.9000006
470+8.50000000,
-4.50000000,
+3.50000000,
-2.50000000,
-1.50000000,
-4.50000000,
-6.50000000,
-4.50000000,
+3.50000000,
+8.50000000
471+5.09999943,
-5.90000057,
-0.900000572,
+5.09999943,
-4.90000057,
+2.09999943,
+7.09999943,
+0.999994278E-1,
+0.999994278E-1,
-7.90000057
473call setCov(cov, uppDia, mean, sample, dim)
475+28.0500011,
-6.15000010
476+0.00000000,
+22.8899994
478call setCov(cov, uppDia, samShifted, dim)
480+28.0500011,
-6.15000010
481+0.00000000,
+22.8899994
484call setCov(cov, lowDia, mean, sample, dim)
486+28.0500011,
+0.00000000
487-6.15000010,
+22.8899994
489call setCov(cov, lowDia, samShifted, dim)
491+28.0500011,
+0.00000000
492-6.15000010,
+22.8899994
494'Compute the sample covariance along the first dimension.'
498call setCov(cov, uppDia, mean,
transpose(sample), dim)
500+28.0500011,
-6.15000010
501+0.00000000,
+22.8899994
503call setCov(cov, uppDia,
transpose(samShifted), dim)
505+28.0500011,
-6.15000010
506+0.00000000,
+22.8899994
509call setCov(cov, lowDia, mean,
transpose(sample), dim)
511+28.0500011,
+0.00000000
512-6.15000010,
+22.8899994
514call setCov(cov, lowDia,
transpose(samShifted), dim)
516+28.0500011,
+0.00000000
517-6.15000010,
+22.8899994
519'Compute the full sample covariance for a pair of time series.'
522call setCov(cov, mean, sample(
1,:), sample(
2,:))
524+28.0500011,
-6.15000010
525-6.15000010,
+22.8899994
527call setCov(cov, samShifted(
1,:), samShifted(
2,:))
529+28.0500011,
-6.15000010
530-6.15000010,
+22.8899994
539ndim
= 2; nsam
= 10; dim
= 2
540sample
= reshape(
getUnifRand(
1,
20, ndim
* nsam), shape
= [ndim, nsam], order
= [
2,
1])
542+7.00000000,
+20.0000000,
+18.0000000,
+14.0000000,
+20.0000000,
+7.00000000,
+10.0000000,
+19.0000000,
+7.00000000,
+6.00000000
543+19.0000000,
+1.00000000,
+4.00000000,
+15.0000000,
+10.0000000,
+8.00000000,
+14.0000000,
+7.00000000,
+5.00000000,
+3.00000000
547+3,
+8,
+2,
+4,
+1,
+7,
+8,
+8,
+8,
+3
548call setMean(mean, sample, dim, iweight, iweisum)
550+12.4615393,
+8.00000000
555+3,
+8,
+2,
+4,
+1,
+7,
+8,
+8,
+8,
+3
556call setMean(mean, sample, dim, rweight, rweisum)
558+12.4615393,
+8.00000000
568-5.46153927,
+7.53846073,
+5.53846073,
+1.53846073,
+7.53846073,
-5.46153927,
-2.46153927,
+6.53846073,
-5.46153927,
-6.46153927
569+11.0000000,
-7.00000000,
-4.00000000,
+7.00000000,
+2.00000000,
+0.00000000,
+6.00000000,
-1.00000000,
-3.00000000,
-5.00000000
571call setCov(cov, uppDia, mean, sample, dim, iweight, iweisum)
573+31.4408321,
-10.2115393
574+0.00000000,
+27.5000019
576call setCov(cov, uppDia, samShifted, dim, iweight,
sum(iweight))
578+31.4408321,
-10.2115393
579+0.00000000,
+27.5000019
582call setCov(cov, lowDia, mean, sample, dim, iweight, iweisum)
584+31.4408321,
+0.00000000
585-10.2115393,
+27.5000019
587call setCov(cov, lowDia, samShifted, dim, iweight,
sum(iweight))
589+31.4408321,
+0.00000000
590-10.2115393,
+27.5000019
592'Compute the sample covariance along the first dimension.'
596call setCov(cov, uppDia, mean,
transpose(sample), dim, iweight, iweisum)
598+31.4408321,
-10.2115393
599+0.00000000,
+27.5000019
601call setCov(cov, uppDia,
transpose(samShifted), dim, iweight,
sum(iweight))
603+31.4408321,
-10.2115393
604+0.00000000,
+27.5000019
607call setCov(cov, lowDia, mean,
transpose(sample), dim, iweight, iweisum)
609+31.4408321,
+0.00000000
610-10.2115393,
+27.5000019
612call setCov(cov, lowDia,
transpose(samShifted), dim, iweight,
sum(iweight))
614+31.4408321,
+0.00000000
615-10.2115393,
+27.5000019
617'Compute the full sample covariance for a pair of time series.'
620call setCov(cov, mean, sample(
1,:), sample(
2,:), iweight, iweisum)
622+31.4408321,
-10.2115393
623-10.2115393,
+27.5000019
625call setCov(cov, samShifted(
1,:), samShifted(
2,:), iweight,
sum(iweight))
627+31.4408321,
-10.2115393
628-10.2115393,
+27.5000019
638-5.46153927,
+7.53846073,
+5.53846073,
+1.53846073,
+7.53846073,
-5.46153927,
-2.46153927,
+6.53846073,
-5.46153927,
-6.46153927
639+11.0000000,
-7.00000000,
-4.00000000,
+7.00000000,
+2.00000000,
+0.00000000,
+6.00000000,
-1.00000000,
-3.00000000,
-5.00000000
641call setCov(cov, uppDia, mean, sample, dim, rweight, rweisum)
643+31.4408321,
-10.2115393
644+0.00000000,
+27.5000019
646call setCov(cov, uppDia, samShifted, dim, rweight,
sum(rweight))
648+31.4408321,
-10.2115393
649+0.00000000,
+27.5000019
652call setCov(cov, lowDia, mean, sample, dim, rweight, rweisum)
654+31.4408321,
+0.00000000
655-10.2115393,
+27.5000019
657call setCov(cov, lowDia, samShifted, dim, rweight,
sum(rweight))
659+31.4408321,
+0.00000000
660-10.2115393,
+27.5000019
662'Compute the sample covariance along the first dimension.'
666call setCov(cov, uppDia, mean,
transpose(sample), dim, rweight, rweisum)
668+31.4408321,
-10.2115393
669+0.00000000,
+27.5000019
671call setCov(cov, uppDia,
transpose(samShifted), dim, rweight,
sum(rweight))
673+31.4408321,
-10.2115393
674+0.00000000,
+27.5000019
677call setCov(cov, lowDia, mean,
transpose(sample), dim, rweight, rweisum)
679+31.4408321,
+0.00000000
680-10.2115393,
+27.5000019
682call setCov(cov, lowDia,
transpose(samShifted), dim, rweight,
sum(rweight))
684+31.4408321,
+0.00000000
685-10.2115393,
+27.5000019
687'Compute the full sample covariance for a pair of time series.'
690call setCov(cov, mean, sample(
1,:), sample(
2,:), rweight, rweisum)
692+31.4408321,
-10.2115393
693-10.2115393,
+27.5000019
695call setCov(cov, samShifted(
1,:), samShifted(
2,:), rweight,
sum(rweight))
697+31.4408321,
-10.2115393
698-10.2115393,
+27.5000019