Return the (weighted) mean of a pair of time series or of an input sample of nsam observations with ndim = 1 or 2 attributes, optionally weighted by the input weight, optionally also sum(weight) and optionally, sum(weight**2).
This generic interface is developed to specifically improve the performance of other procedures where sum(weight) or sum(weight**2) are also needed in addition to the mean of the weighted sample.
For example, this situation occurs frequently in the computation of the variance or covariance matrix of a weighted sample.
- Parameters
-
| [out] | mean | : The output object of,
-
type
complex of kind any supported by the processor (e.g., CK, CK32, CK64, or CK128),
-
type
real of kind any supported by the processor (e.g., RK, RK32, RK64, or RK128),
containing the data mean.
-
When the input
sample has the shape (nsam), the output mean must be a scalar.
-
When the input
x and y are present, the output mean must be of shape mean(1:2).
-
When the input
sample has the shape (nsam, ndim) or (ndim, nsam), the output mean must be a vector of length ndim.
|
| [in] | x | : The contiguous array of shape (nsam) of the same type and kind as the output mean, containing the first of the two data series whose mean is to be computed.
(optional. It must be present *if and only if** the input argument sample is missing and y is present.) |
| [in] | y | : The contiguous of the same type and kind and shape as the input x, containing the second of the two data series whose mean is to be computed.
(optional. It must be present *if and only if** the input argument sample is missing and x is present.) |
| [in] | sample | : The contiguous array of shape (nsam), (ndim, nsam) or (nsam, ndim) of the same type and kind as the output mean, containing the sample whose mean is to be computed.
(optional. It must be present *if and only if** the input arguments x and y are missing.) |
| [in] | dim | : The input scalar integer of default kind IK representing the dimension (1 or 2) of the input sample along which the mean must be computed.
-
If
dim = 1, the input sample of rank 2 is assumed to have the shape (nsam, ndim).
-
If
dim = 2, the input sample of rank 2 is assumed to have the shape (ndim, nsam).
The input dim must always be 1 or missing for an input sample of rank 1.
(optional. It can be present only if sample is also present. If missing, the mean of the whole input sample is computed.) |
| [in] | weight | : The contiguous vector of length nsam of
-
type
integer of default kind IK, or
-
type
real of the same kind as that of the output mean,
containing the corresponding weights of the data points in the input sample or the input pair x and y.
|
| [out] | weisum | : The output scalar of the same type and kind as the input weight.
On output, it will contain sum(weight).
This quantity is frequently needed in computing the weighted sample variance.
(optional. It must be present if and only if the input argument weight is also present.) |
Possible calling interfaces ⛓
call setMean(mean(
1:
2), x(
1 : nsam), y(
1 : nsam))
call setMean(mean(
1:
2), x(
1 : nsam), y(
1 : nsam), weight(
1 : nsam), weisum)
call setMean(mean, sample(
1 : nsam))
call setMean(mean, sample(
1 : nsam), weight(
1 : nsam), weisum)
call setMean(mean, sample(
1 : nsam), dim)
call setMean(mean, sample(
1 : nsam), dim, weight(
1 : nsam), weisum)
call setMean(mean(
1 : ndim), sample(:,:))
call setMean(mean(
1 : ndim), sample(:,:), weight(
1 : nsam), weisum)
call setMean(mean(
1 : ndim), sample(:,:), dim)
call setMean(mean(
1 : ndim), sample(:,:), dim, weight(
1 : nsam), weisum)
Return the (weighted) mean of a pair of time series or of an input sample of nsam observations with n...
This module contains classes and procedures for computing the first moment (i.e., the statistical mea...
- Warning
- The condition
all(0. <= weight) must hold for the corresponding input arguments.
The condition 1 <= dim .and. dim <= rank(sample) must hold for the corresponding input arguments.
The condition size(sample, dim) == size(weight, 1) must hold for the corresponding input arguments.
The condition (dim == 1 .and. size(mean, 1) == size(sample, 2)) .or. (dim == 2 .and. size(mean, 1) == size(sample, 1)) must hold for the corresponding input arguments.
The condition size(x) == size(weight) must hold for the corresponding input arguments.
The condition size(x) == size(y) must hold for the corresponding input arguments.
These conditions are verified only if the library is built with the preprocessor macro CHECK_ENABLED=1.
-
The
pure procedure(s) documented herein become impure when the ParaMonte library is compiled with preprocessor macro CHECK_ENABLED=1.
By default, these procedures are pure in release build and impure in debug and testing builds.
- Note
- If the input sample is to be an array of type
integer, simply convert the sample to an array of type real of the desired kind for the output real mean of the sample.
There is no point in accepting an input sample of type integer since it will be inevitably converted to an array of type real within the procedure to avoid potential integer overflow.
Furthermore, an sample of type integer creates ambiguity about the kind of the real-valued returned mean by the procedure.
See the notes in the description of the pm_sampleMean.
-
Note that the mean of any one or two-dimensional sample can be simply computed via the Fortran intrinsic routine
sum(): integer :: i
integer , parameter :: NDIM = 3_IK
integer , parameter :: NSAM = 1000_IK
real , parameter :: sample(NDIM,NSAM) = reshape([( real(i,RK), i = 1, NSAM )], shape = shape(sample))
real , allocatable :: mean(:)
mean = sum(sample, dim = 1) / size(transpose(sample), dim = 1)
mean = sum(sample, dim = 2) / size(sample, dim = 2) ! assuming the second dimension represents the observations.
-
The mean of a whole multidimensional array can be obtained by either,
-
reshaping the array to a vector form and passing it to this procedure, or
-
mapping the array to a 1-dimensional pointer array of the same size as the
ndim dimensional array.
See the examples below.
- Developer Remark:
- The logic behind allowing a pair of time series data
x and y is to allow fast computation of the mean of the pair and the weight sums in one loop.
This pattern occurs frequently in the computation of correlation coefficients.
- See also
- getVar
getMean
setVarMean
Example usage ⛓
12 integer(IK) :: idim, ndim, nsam
13 type(display_type) :: disp
17 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
18 call disp%show(
"!Compute the mean of a 1-D array.")
19 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
24 real(TKG),
allocatable :: sample(:)
26 call disp%show(
"nsam = getUnifRand(1, 5)")
28 call disp%show(
"sample = getUnifRand(0., 1., nsam)")
32 call disp%show(
"call setMean(mean, sample)")
36 call disp%show(
"call setMean(mean, sample, dim = 1_IK)")
37 call setMean(mean, sample,
dim = 1_IK)
44 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
45 call disp%show(
"!Compute the mean of a pair of data series.")
46 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
51 real(TKG),
allocatable :: sample(:,:)
52 real(TKG),
allocatable :: weight(:)
55 call disp%show(
"nsam = getUnifRand(1, 5)")
57 call disp%show(
"sample = getUnifRand(0., 1., nsam, 2_IK)")
61 call disp%show(
"call setMean(mean, sample(:,1), sample(:,2))")
62 call setMean(mean, sample(:,
1), sample(:,
2))
65 call disp%show(
"call setMean(mean, sample, dim = 1_IK) ! for comparison.")
66 call setMean(mean, sample,
dim = 1_IK)
70 call disp%show(
"weight = getUnifRand(1, 100, nsam)")
74 call disp%show(
"call setMean(mean, sample(:,1), sample(:,2), weight, weisum)")
75 call setMean(mean, sample(:,
1), sample(:,
2), weight, weisum)
80 call disp%show(
"call setMean(mean, sample, 1_IK, weight, weisum) ! for comparison.")
81 call setMean(mean, sample,
1_IK, weight, weisum)
87 call disp%show(
"call setMean(mean, sample(:,1), sample(:,2), weight, weisum)")
88 call setMean(mean, sample(:,
1), sample(:,
2), weight, weisum)
93 call disp%show(
"call setMean(mean, sample, 1_IK, weight, weisum) ! for comparison.")
94 call setMean(mean, sample,
1_IK, weight, weisum)
103 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
104 call disp%show(
"!Compute the mean of a 2-D array along a specific dimension.")
105 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
109 real(TKG),
allocatable :: mean(:)
110 real(TKG),
allocatable :: sample(:,:)
112 call disp%show(
"ndim = getUnifRand(1, 3); nsam = getUnifRand(1, 5)")
114 call disp%show(
"sample = getUnifRand(0., 1., ndim, nsam)")
118 call disp%show(
"call setResized(mean, ndim)")
120 call disp%show(
"call setMean(mean(1), sample)")
124 call disp%show(
"call setMean(mean, sample, dim = 2_IK)")
125 call setMean(mean, sample,
dim = 2_IK)
128 call disp%show(
"call setMean(mean, transpose(sample), dim = 1_IK)")
129 call setMean(mean,
transpose(sample),
dim = 1_IK)
136 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
137 call disp%show(
"!Compute the mean of a 1-D weighted array.")
138 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
143 real(TKG),
allocatable :: sample(:)
144 real(TKG),
allocatable :: weight(:)
147 call disp%show(
"nsam = getUnifRand(1, 5)")
149 call disp%show(
"sample = getUnifRand(0., 1., nsam)")
153 call disp%show(
"weight = getUnifRand(1, 100, nsam)")
157 call disp%show(
"call setMean(mean, sample)")
161 call disp%show(
"call setMean(mean, sample, dim = 1_IK)")
162 call setMean(mean, sample,
dim = 1_IK)
165 call disp%show(
"call setMean(mean, sample, weight, weisum)")
166 call setMean(mean, sample, weight, weisum)
167 call disp%show(
"[weisum, sum(weight)]")
168 call disp%show( [weisum,
sum(weight)] )
171 call disp%show(
"call setMean(mean, sample, 1_IK, weight, weisum)")
172 call setMean(mean, sample,
1_IK, weight, weisum)
173 call disp%show(
"[weisum, sum(weight)]")
174 call disp%show( [weisum,
sum(weight)] )
181 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
182 call disp%show(
"!Compute the mean of a 2-D weighted array along a specific dimension.")
183 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
187 real(TKG),
allocatable :: mean(:)
188 real(TKG),
allocatable :: sample(:,:)
189 real(TKG),
allocatable :: weight(:)
192 call disp%show(
"ndim = getUnifRand(1, 3); nsam = getUnifRand(1, 5)")
194 call disp%show(
"sample = getUnifRand(0., 1., ndim, nsam)")
198 call disp%show(
"weight = getUnifRand(1, 100, nsam)")
202 call disp%show(
"call setResized(mean, ndim)")
204 call disp%show(
"call setMean(mean(1), sample, [(weight, idim = 1, ndim)], weisum)")
205 call setMean(mean(
1), sample, [(weight, idim
= 1, ndim)], weisum)
208 call disp%show(
"call setMean(mean, sample, 2_IK, weight, weisum)")
209 call setMean(mean, sample,
2_IK, weight, weisum)
210 call disp%show(
"[weisum, sum(weight)]")
211 call disp%show( [weisum,
sum(weight)] )
214 call disp%show(
"call setMean(mean, transpose(sample), 1_IK, weight, weisum)")
215 call setMean(mean,
transpose(sample),
1_IK, weight, weisum)
216 call disp%show(
"[weisum, sum(weight)]")
217 call disp%show( [weisum,
sum(weight)] )
224 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
225 call disp%show(
"!Compute the mean of a multidimensional array by associating it with a 1D pointer.")
226 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
231 integer(IK) :: nslice
232 real(TKG),
allocatable,
target :: sample(:,:,:)
233 real(TKG),
pointer :: samptr(:)
235 call disp%show(
"ndim = getUnifRand(1, 2); nsam = getUnifRand(1, 3); nslice = getUnifRand(1, 4)")
237 call disp%show(
"sample = getUnifRand(0., 1., ndim, nsam, nslice)")
243 call disp%show(
"samptr(1:product(shape(sample))) => sample")
244 samptr(
1:
product(
shape(sample)))
=> sample
245 call disp%show(
"call setMean(mean, samptr)")
251 call disp%show(
"call setMean(mean, reshape(sample, [product(shape(sample))]))")
252 call setMean(mean,
reshape(sample, [
product(
shape(sample))]))
Allocate or resize (shrink or expand) an input allocatable scalar string or array of rank 1....
Generate and return a scalar or a contiguous array of rank 1 of length s1 of randomly uniformly distr...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
This module contains procedures and generic interfaces for resizing allocatable arrays of various typ...
This module contains classes and procedures for computing various statistical quantities related to t...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
integer, parameter RKS
The single-precision real kind in Fortran mode. On most platforms, this is an 32-bit real kind.
Generate and return an object of type display_type.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example output ⛓
10+0.318599999,
+0.544942737,
+0.330364645,
+0.672035933
14call setMean(mean, sample,
dim = 1_IK)
27+0.718679965,
+0.958418965
28+0.973525763,
+0.695811570
29+0.675242543E-1,
+0.360647440
30+0.804494441,
+0.568368971
31call setMean(mean, sample(:,
1), sample(:,
2))
33+0.641056061,
+0.645811737
34call setMean(mean, sample,
dim = 1_IK)
36+0.641056061,
+0.645811737
40+18.0000000,
+27.0000000,
+90.0000000,
+22.0000000
41call setMean(mean, sample(:,
1), sample(:,
2), weight, weisum)
45+0.401257932,
+0.515928924
46call setMean(mean, sample,
1_IK, weight, weisum)
50+0.401257962,
+0.515928924
52call setMean(mean, sample(:,
1), sample(:,
2), weight, weisum)
56+0.401257932,
+0.515928924
57call setMean(mean, sample,
1_IK, weight, weisum)
61+0.401257962,
+0.515928924
72+0.123162985,
+0.306564033,
+0.823826253,
+0.426583886,
+0.964393020
73+0.272504985,
+0.144482970,
+0.876074016,
+0.404005587,
+0.852061808
78call setMean(mean, sample,
dim = 2_IK)
80+0.528906047,
+0.509825885
81call setMean(mean,
transpose(sample),
dim = 1_IK)
83+0.528906047,
+0.509825885
94+0.346967459,
+0.573612690,
+0.913797557,
+0.561539531
97+66.0000000,
+7.00000000,
+87.0000000,
+18.0000000
101call setMean(mean, sample,
dim = 1_IK)
104call setMean(mean, sample, weight, weisum)
106+178.000000,
+178.000000
109call setMean(mean, sample,
1_IK, weight, weisum)
111+178.000000,
+178.000000
129call setMean(mean(
1), sample, [(weight, idim
= 1, ndim)], weisum)
132call setMean(mean, sample,
2_IK, weight, weisum)
134+12.0000000,
+12.0000000
137call setMean(mean,
transpose(sample),
1_IK, weight, weisum)
139+12.0000000,
+12.0000000
153+0.670179784,
+0.769957662,
+0.673075318
155+0.790455043,
+0.705637157,
+0.398651958
157+0.320507646,
+0.898947775,
+0.556919992
159+0.903167963,
+0.722648501,
+0.856229603
162samptr(
1:
product(
shape(sample)))
=> sample
167call setMean(mean,
reshape(sample, [
product(
shape(sample))]))
- Benchmarks:
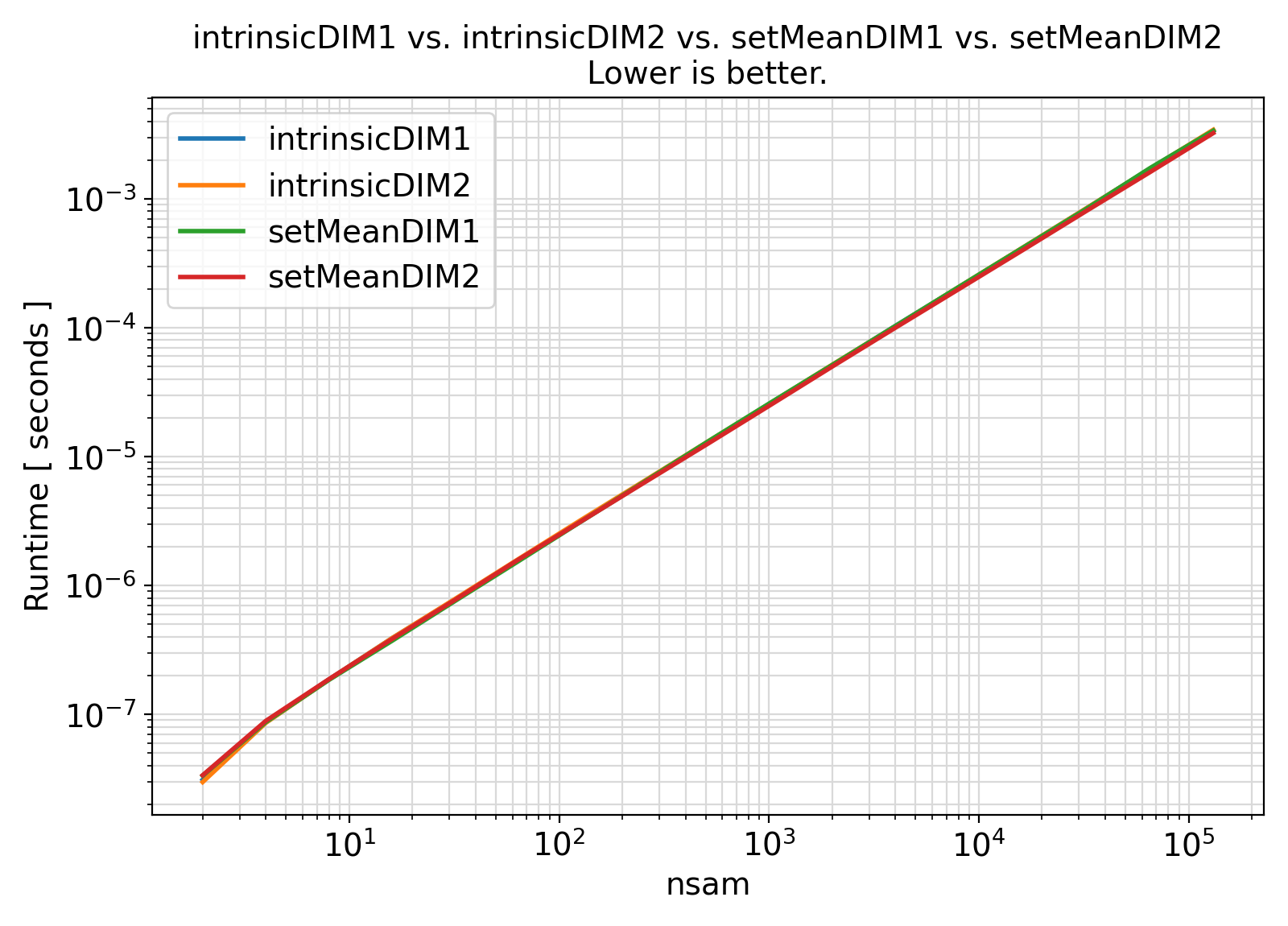
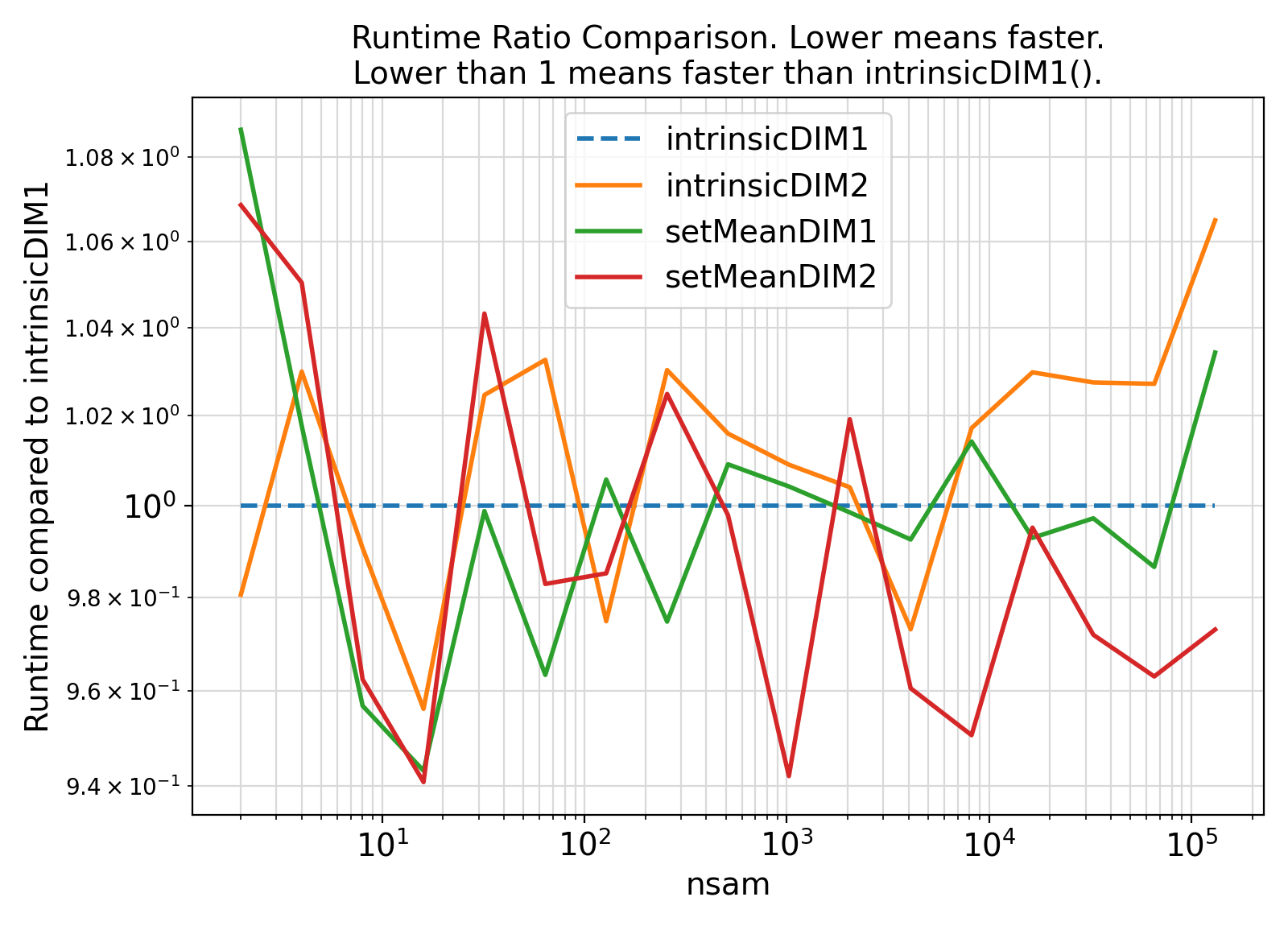
Benchmark :: The runtime performance of setMean along different sample dimensions. ⛓
9 integer(IK) :: itry, ntry
12 integer(IK) :: fileUnit
13 integer(IK) ,
parameter :: NARR
= 18_IK
14 real(RKG) ,
allocatable :: samdim1(:,:)
15 real(RKG) ,
allocatable :: samdim2(:,:)
16 type(bench_type),
allocatable :: bench(:)
17 integer(IK) ,
parameter :: nsammax
= 2**NARR
18 integer(IK) ,
parameter :: ndim
= 5_IK
19 real(RKG) :: mean(ndim)
20 integer(IK) :: isam, nsam
23 bench
= [
bench_type(name
= SK_
"intrinsicDIM1", exec
= intrinsicDIM1, overhead
= setOverhead)
&
24 ,
bench_type(name
= SK_
"intrinsicDIM2", exec
= intrinsicDIM2, overhead
= setOverhead)
&
25 ,
bench_type(name
= SK_
"setMeanDIM1", exec
= setMeanDIM1, overhead
= setOverhead)
&
26 ,
bench_type(name
= SK_
"setMeanDIM2", exec
= setMeanDIM2, overhead
= setOverhead)
&
29 write(
*,
"(*(g0,:,' '))")
30 write(
*,
"(*(g0,:,' '))")
"sample mean benchmarking..."
31 write(
*,
"(*(g0,:,' '))")
33 open(newunit
= fileUnit, file
= "main.out", status
= "replace")
35 write(fileUnit,
"(*(g0,:,','))")
"nsam", (bench(i)
%name, i
= 1,
size(bench))
38 loopOverMatrixSize:
do iarr
= 1, NARR
- 1
42 allocate(samdim1(nsam, ndim), samdim2(ndim, nsam))
43 write(
*,
"(*(g0,:,' '))")
"Benchmarking setMeanDIM1() vs. setMeanDIM2()", nsam, ntry
49 write(fileUnit,
"(*(g0,:,','))") nsam, (bench(i)
%timing
%mean
/ ntry, i
= 1,
size(bench))
50 deallocate(samdim1, samdim2)
52 end do loopOverMatrixSize
53 write(
*,
"(*(g0,:,' '))") dumm
63 subroutine setOverhead()
65 call random_number(mean)
70 subroutine setSamDIM1()
72 call random_number(samdim1(isam,
1 : ndim))
76 subroutine setSamDIM2()
78 call random_number(samdim2(
1 : ndim, isam))
82 subroutine setMeanDIM1()
86 call setMean(mean, samdim1,
dim = 1_IK)
91 subroutine setMeanDIM2()
95 call setMean(mean, samdim2,
dim = 2_IK)
100 subroutine intrinsicDIM1()
103 mean
= sum(samdim1,
dim = 1)
/ size(samdim1,
dim = 1)
104 dumm
= dumm
+ mean(
1)
108 subroutine intrinsicDIM2()
111 mean
= sum(samdim2,
dim = 2)
/ size(samdim1,
dim = 2)
112 dumm
= dumm
+ mean(
1)
Generate and return an object of type timing_type containing the benchmark timing information and sta...
This module contains abstract interfaces and types that facilitate benchmarking of different procedur...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter RKD
The double precision real kind in Fortran mode. On most platforms, this is an 64-bit real kind.
This is the class for creating benchmark and performance-profiling objects.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Postprocessing of the benchmark output ⛓
3import matplotlib.pyplot
as plt
8dirname = os.path.basename(os.getcwd())
12df = pd.read_csv(
"main.out", delimiter =
",")
13colnames = list(df.columns.values)
19ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
22for colname
in colnames[1:]:
23 plt.plot( df[colnames[0]].values
28plt.xticks(fontsize = fontsize)
29plt.yticks(fontsize = fontsize)
30ax.set_xlabel(colnames[0], fontsize = fontsize)
31ax.set_ylabel(
"Runtime [ seconds ]", fontsize = fontsize)
32ax.set_title(
" vs. ".join(colnames[1:])+
"\nLower is better.", fontsize = fontsize)
36plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
37ax.tick_params(axis =
"y", which =
"minor")
38ax.tick_params(axis =
"x", which =
"minor")
39ax.legend ( colnames[1:]
46plt.savefig(
"benchmark." + dirname +
".runtime.png")
52ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
55plt.plot( df[colnames[0]].values
56 , np.ones(len(df[colnames[0]].values))
61for colname
in colnames[2:]:
62 plt.plot( df[colnames[0]].values
63 , df[colname].values / df[colnames[1]].values
67plt.xticks(fontsize = fontsize)
68plt.yticks(fontsize = fontsize)
69ax.set_xlabel(colnames[0], fontsize = fontsize)
70ax.set_ylabel(
"Runtime compared to {}".format(colnames[1]), fontsize = fontsize)
71ax.set_title(
"Runtime Ratio Comparison. Lower means faster.\nLower than 1 means faster than {}().".format(colnames[1]), fontsize = fontsize)
75plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
76ax.tick_params(axis =
"y", which =
"minor")
77ax.tick_params(axis =
"x", which =
"minor")
78ax.legend ( colnames[1:]
85plt.savefig(
"benchmark." + dirname +
".runtime.ratio.png")
Visualization of the benchmark output ⛓
Benchmark moral ⛓
- The procedures under the generic interface setMean can compute the covariance under two different sample axes.
- Recall that C is a row-major language while Fortran is a column-major language.
- As such, one would expect the computations for a sample whose observations are stored along the second axis would be faster in the Fortran programming language.
- However, such an expectation does not appear to hold at all times and appears to depend significantly on the computing architecture and the number of data attributes involved.
- The higher the number of data attributes, the more likely the computations along the second axis of
sample will be faster.
- Note that for small number of data attributes, the computations along the second data axis involve a small loop that has significant computational cost due to the implicit branching involved in the loop.
- Test:
- test_pm_sampleMean
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, April 21, 2017, 1:54 AM, Institute for Computational Engineering and Sciences (ICES), The University of Texas Austin
Definition at line 2008 of file pm_sampleMean.F90.