|
ParaMonte MATLAB 3.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
|
ParaMonte MATLAB 3.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
This is the base class for generating instances of objects that contain the specifications of various types of figures.
More...
Public Member Functions | |
| function | Plot (in subplot, in varargin) |
| function | reset (in self, in varargin) |
| Reset the properties of the plot to the original default settings. More... | |
| function | make (in self, in varargin) |
| Configure the plot settings and specifications, make the plot, and return nothing. More... | |
| function | premake (in self, in varargin) |
| Configure the plot settings and specifications and return nothing. More... | |
Data Fields | |
| Property | subplot |
This is the base class for generating instances of objects that contain the specifications of various types of figures.
This is a generic class for generating figures containing a single subplot (axes).
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
Amir Shahmoradi, July 5 2024, 1:07 PM, NASA Goddard Space Flight Center (GSFC), Washington, D.C.
| function Plot::Plot | ( | in | subplot, |
| in | varargin | ||
| ) |
| [in] | subplot | : The input scalar object of superclass pm.vis.Subplot. The input subplot object must minimally have the make() and reset() methods. |
| [in] | varargin | : Any property, value pair of the parent object.If the property is a struct(), then its value must be given as a cell array, with consecutive elements representing the struct property-name, property-value pairs.Note that all of these property-value pairs can be also directly set via the parent object attributes, before calling the make() method.The input varargin can also contain the components of the subplot component of the parent object. |
self : The output scalar object of class pm.vis.Plot.
Possible calling interfaces ⛓
varargin can also contain the components of the subplot component of the parent object.
Example usage ⛓
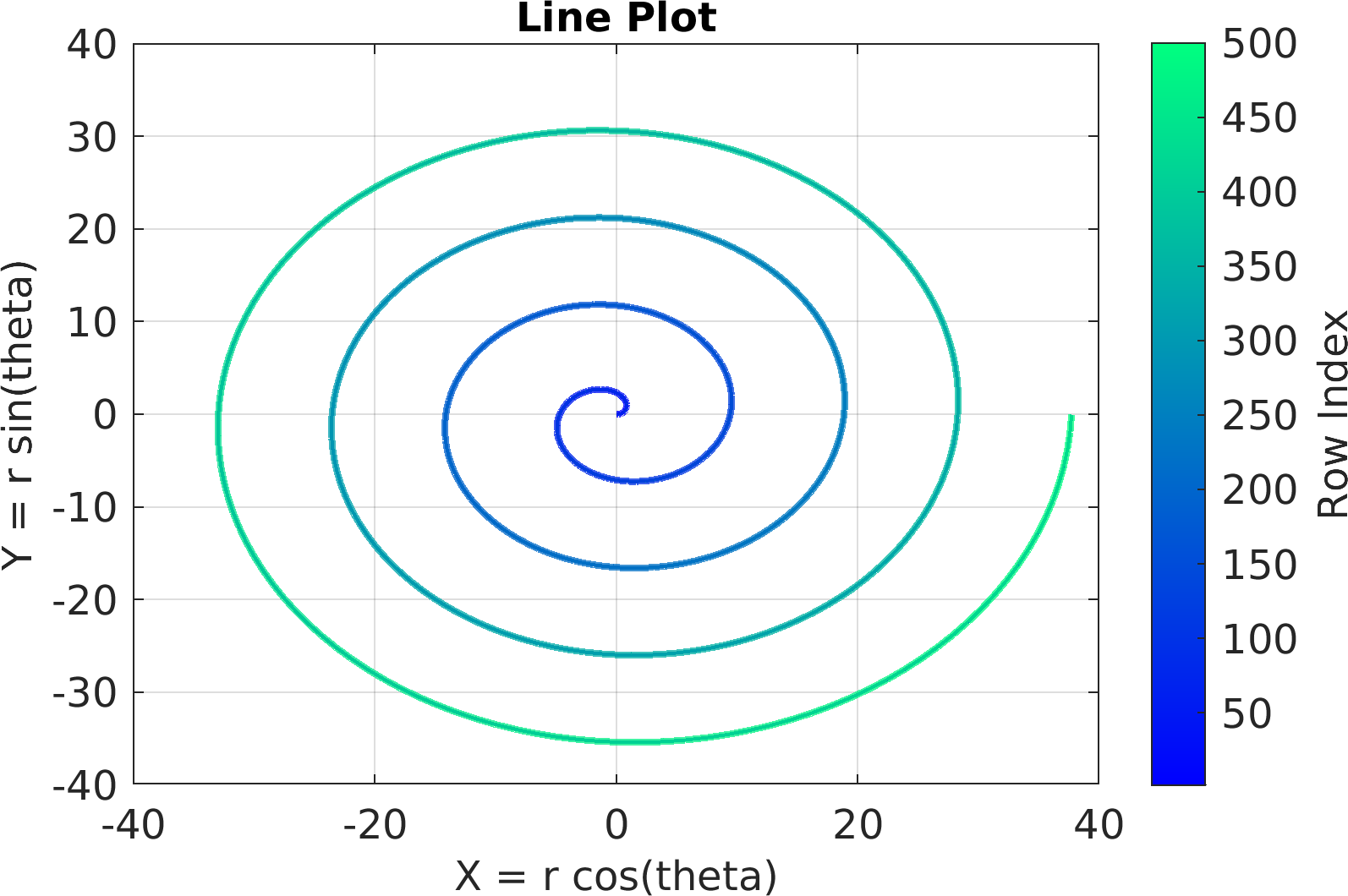
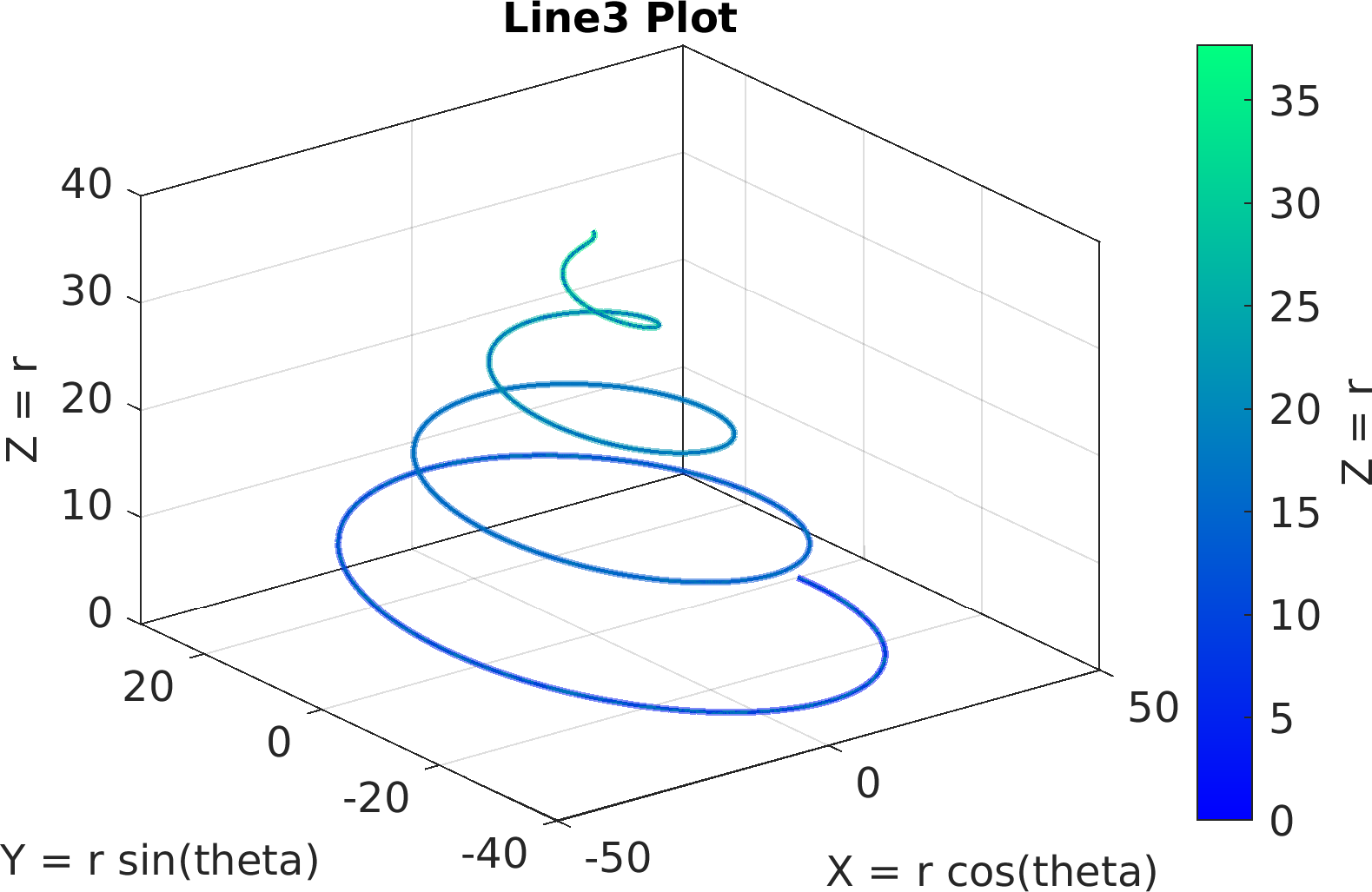
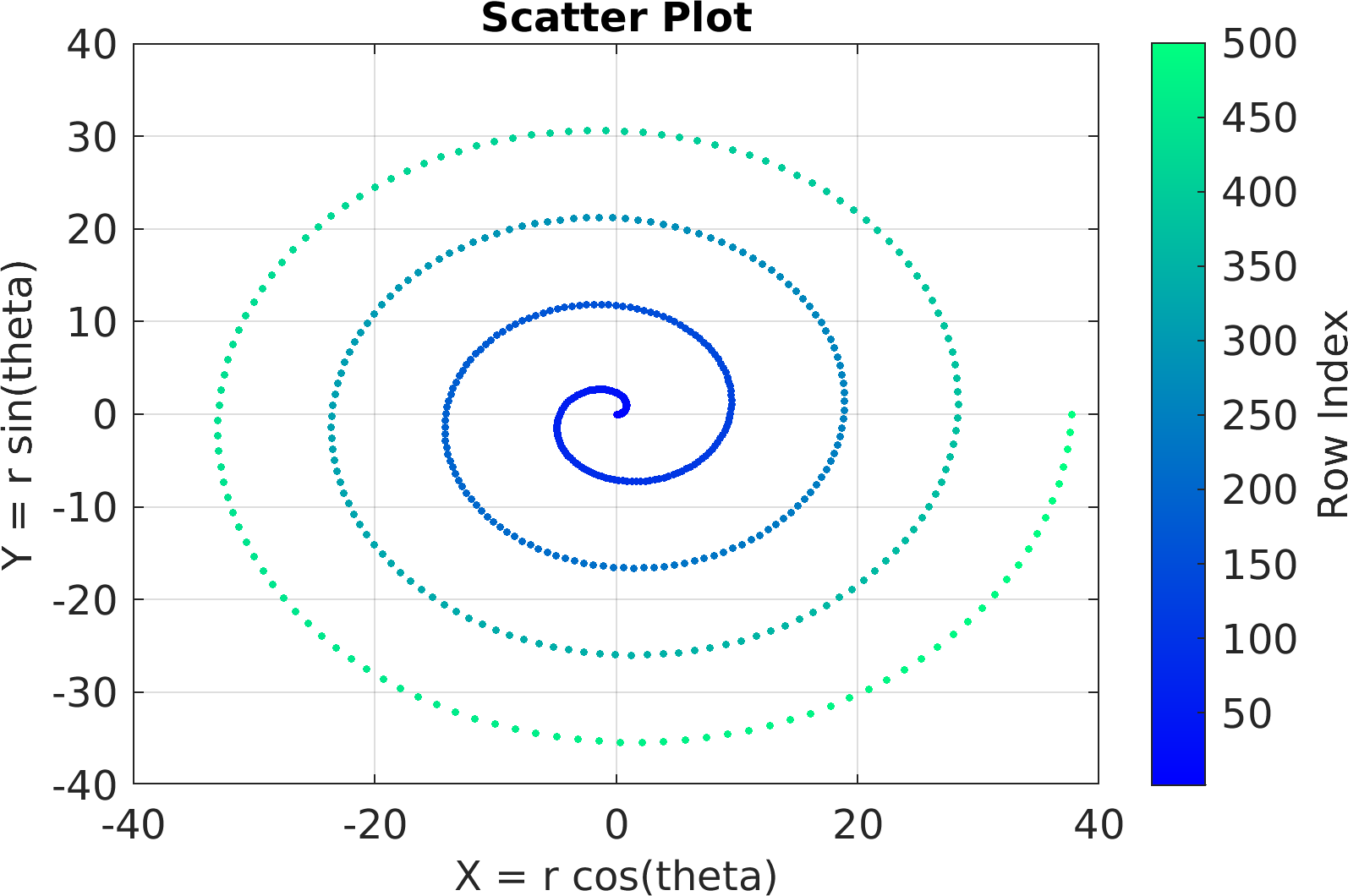
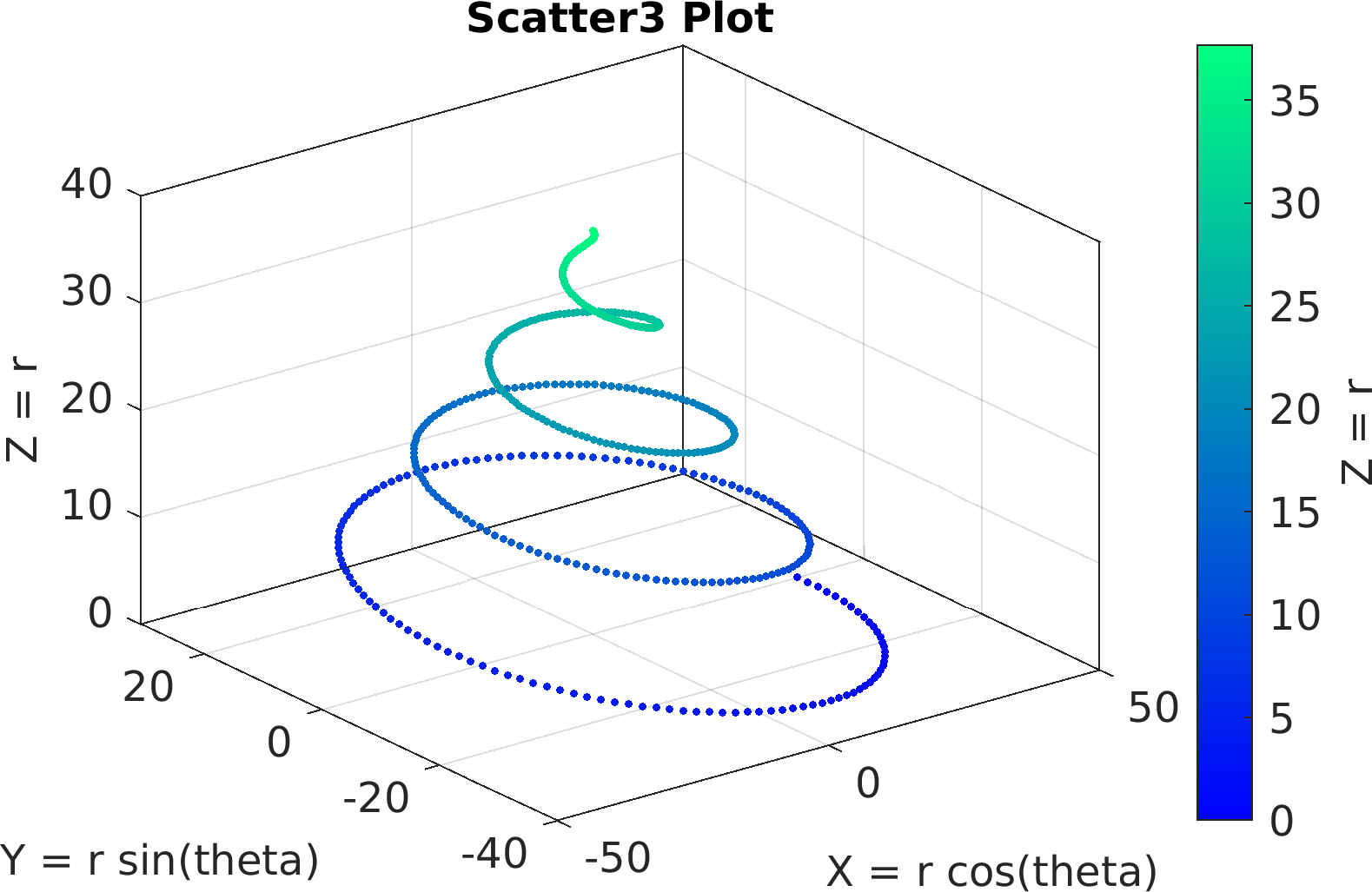
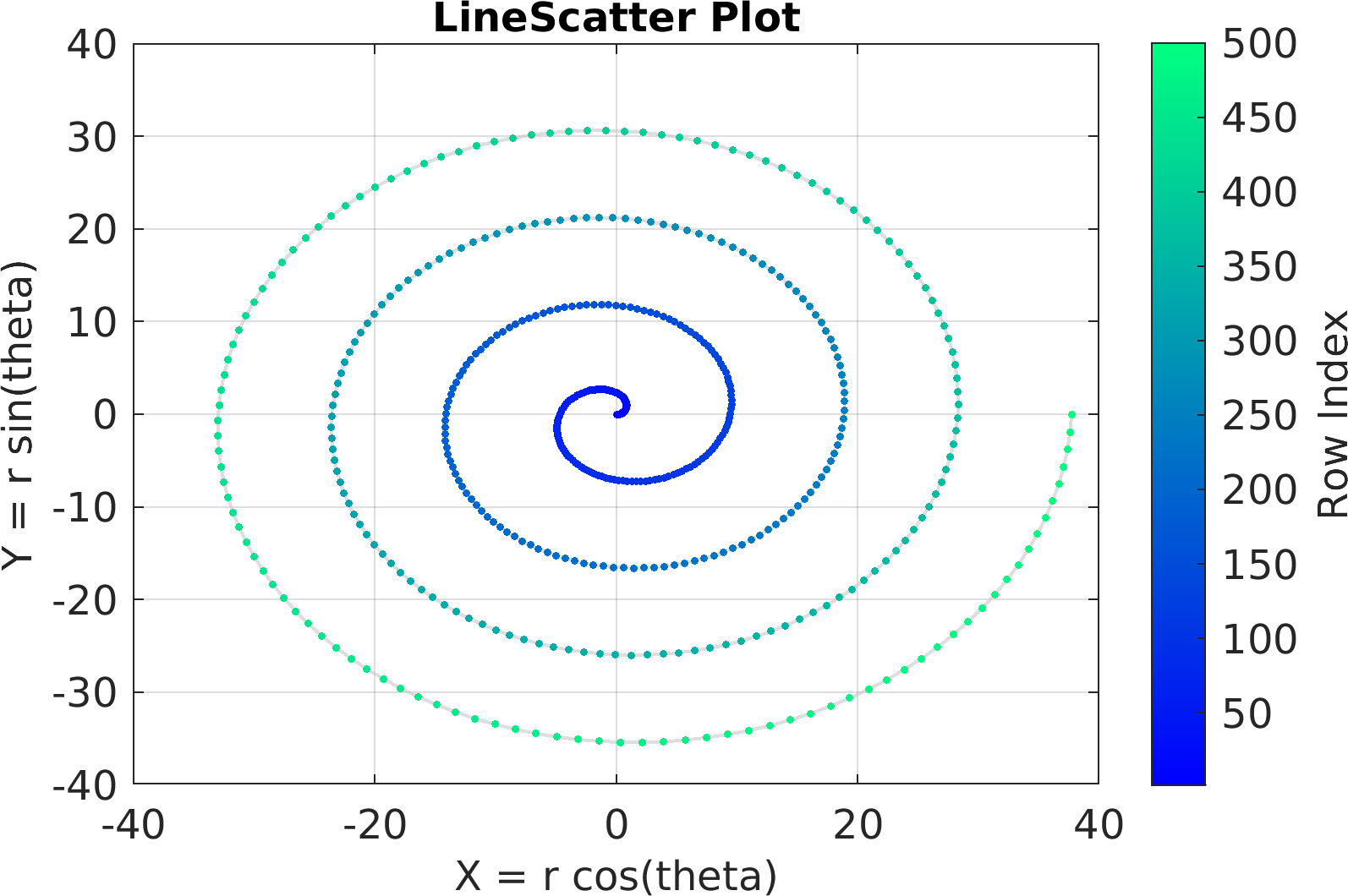
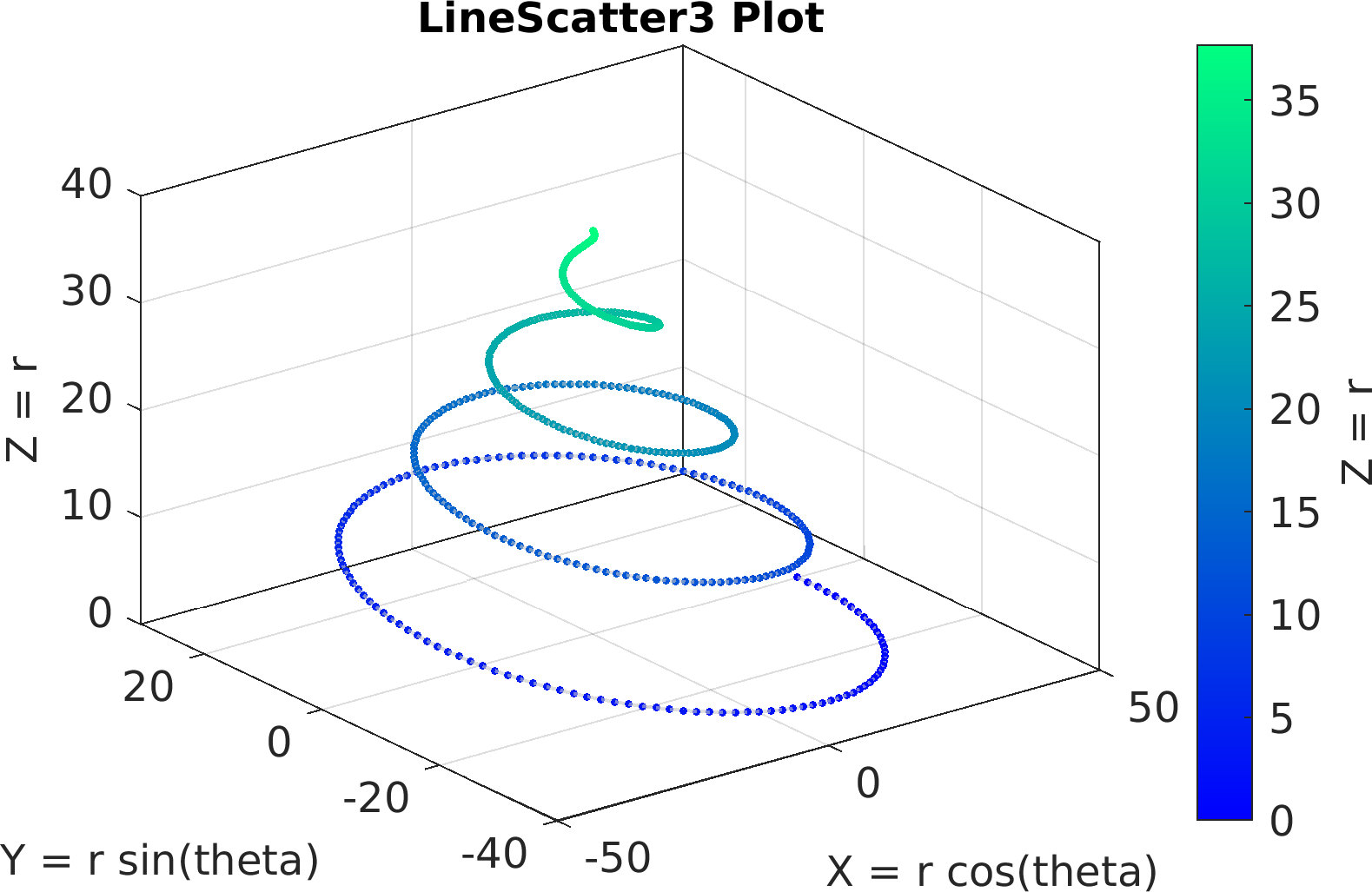
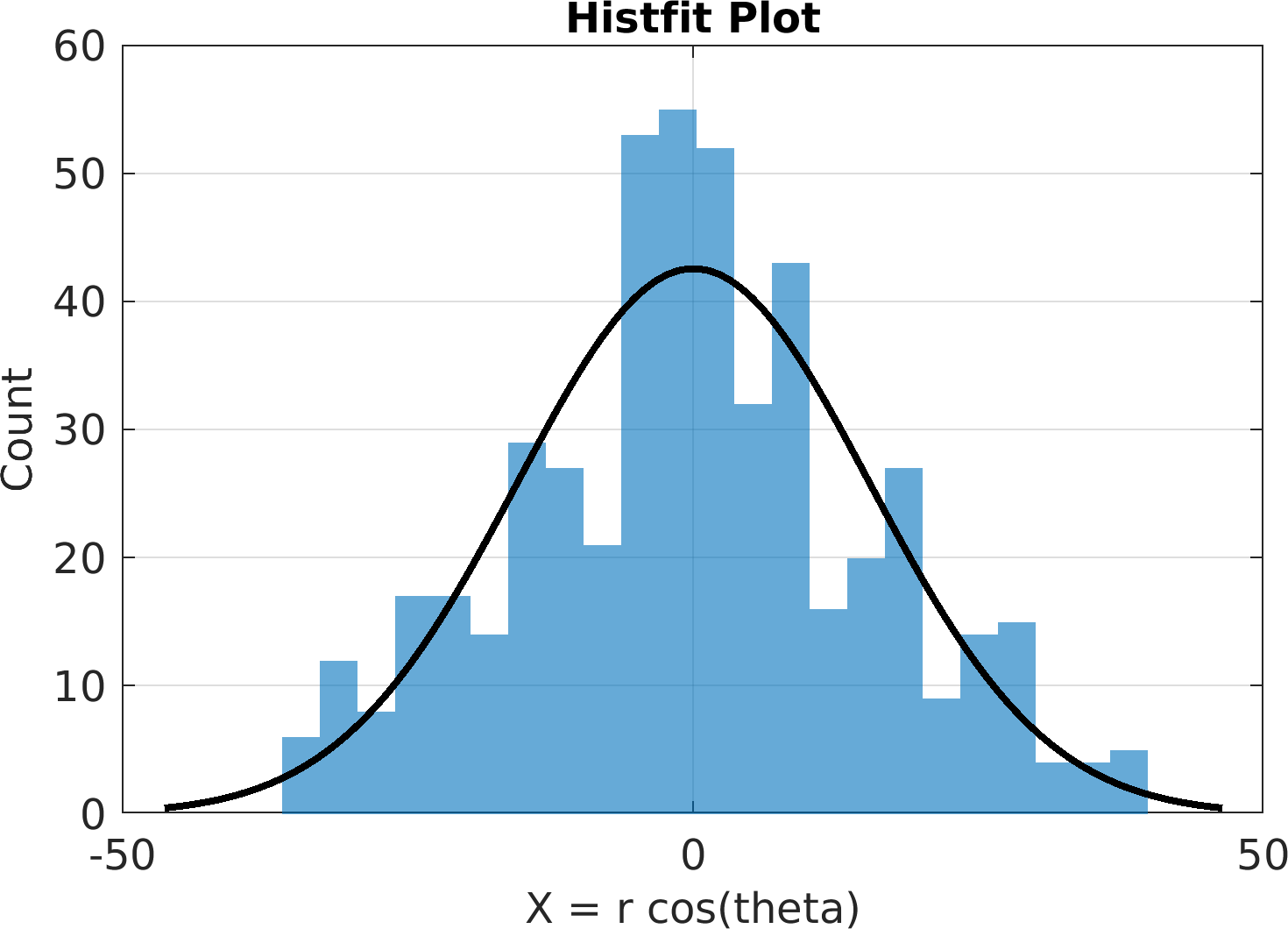
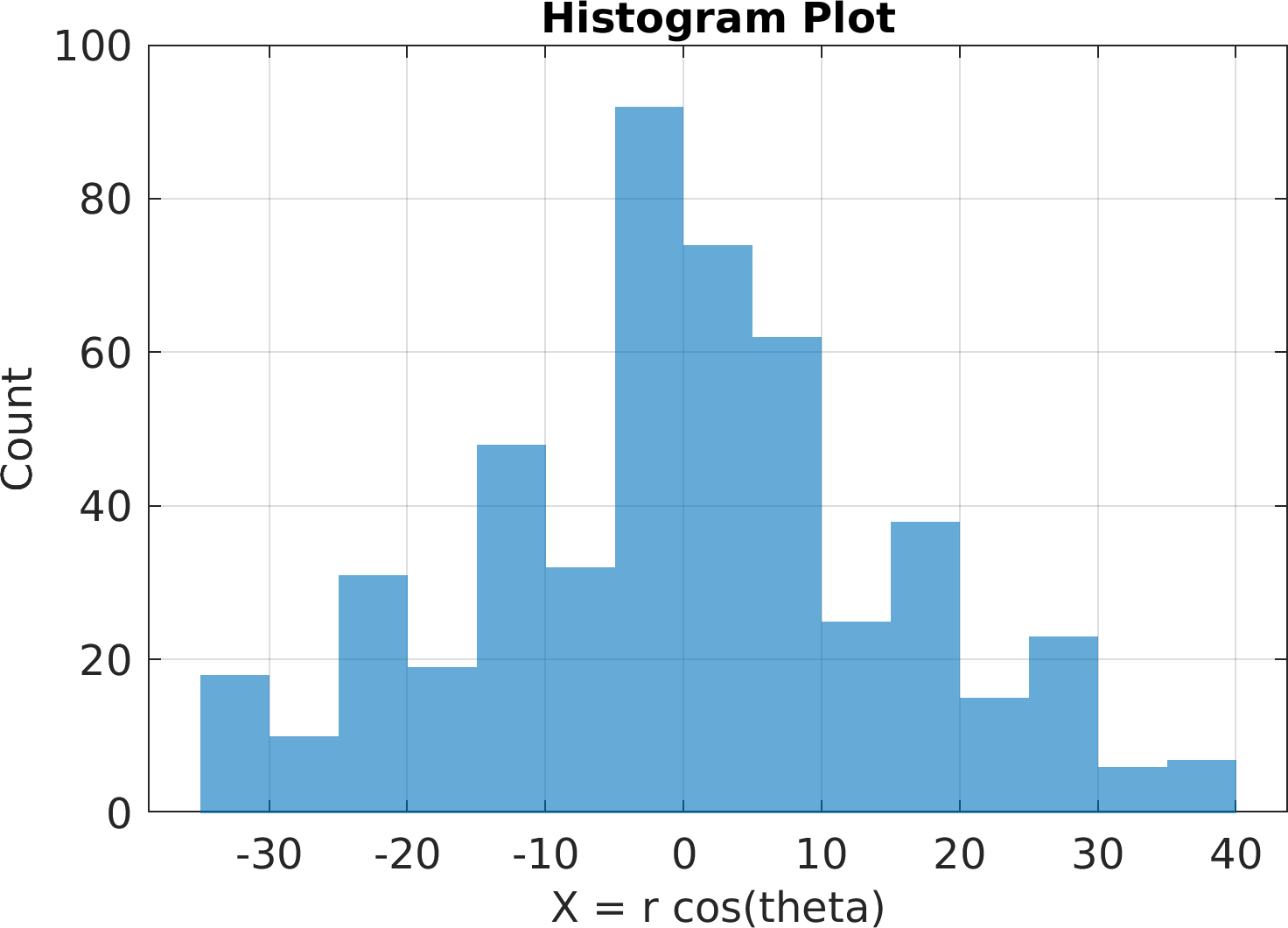
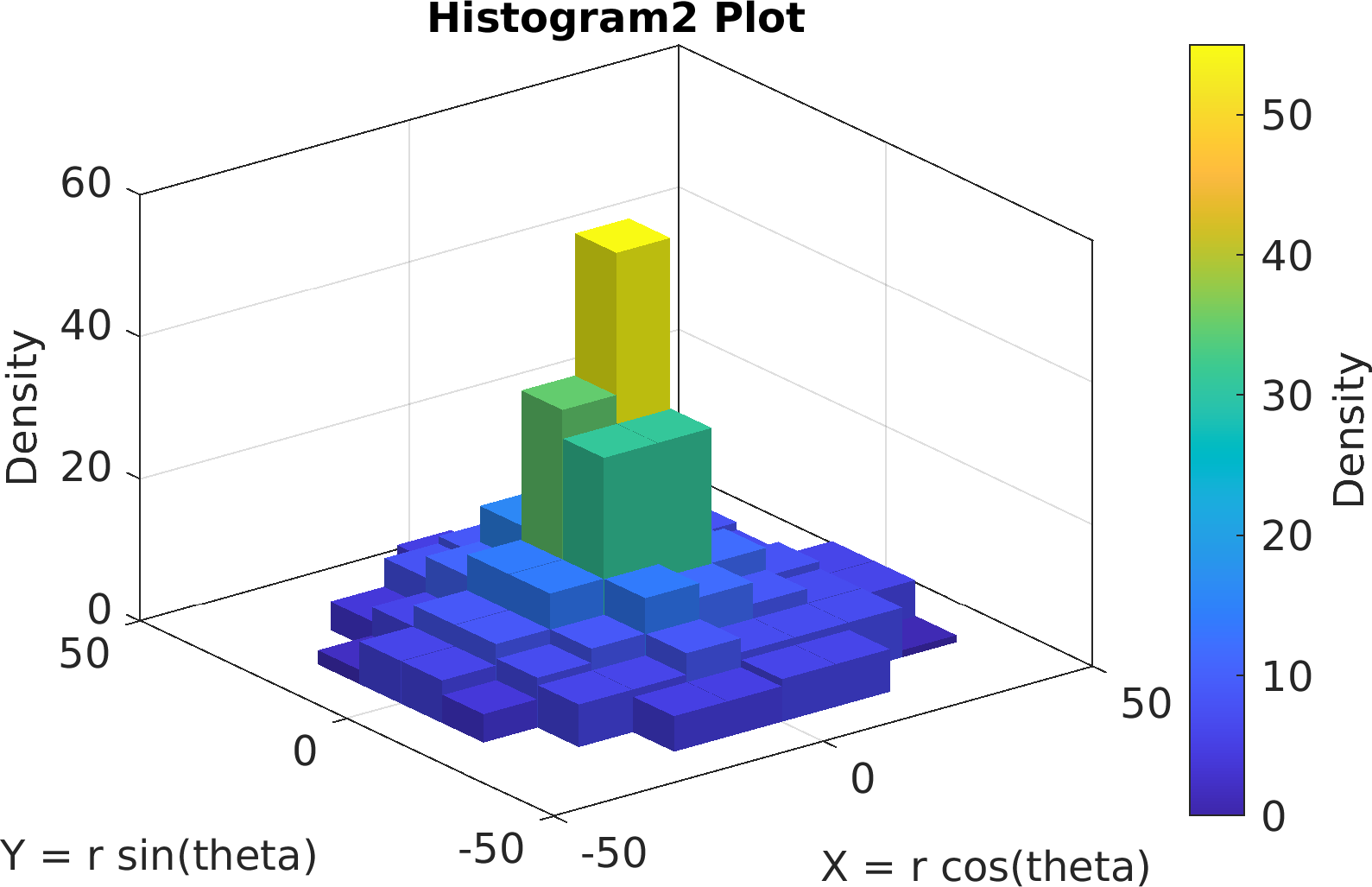
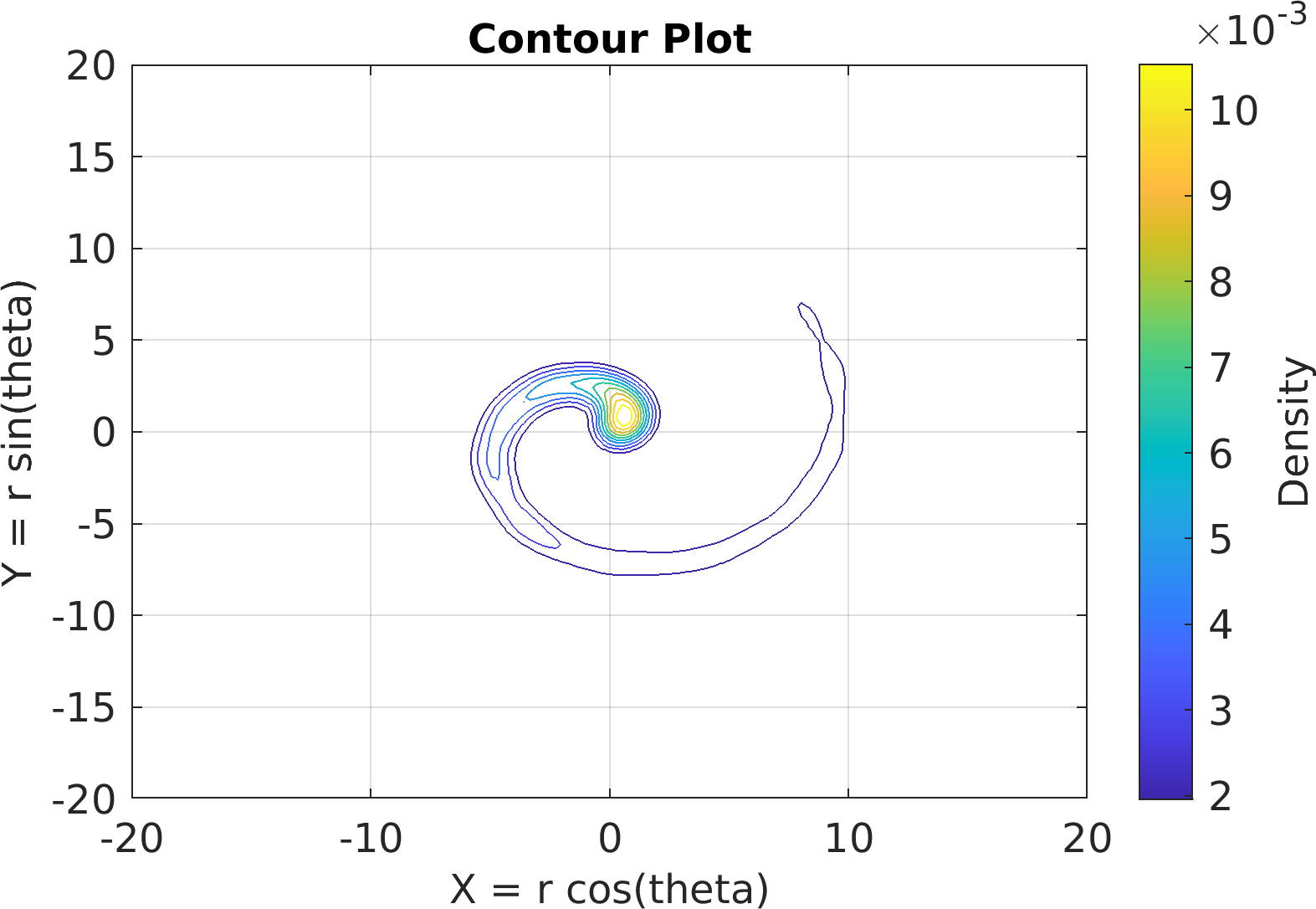
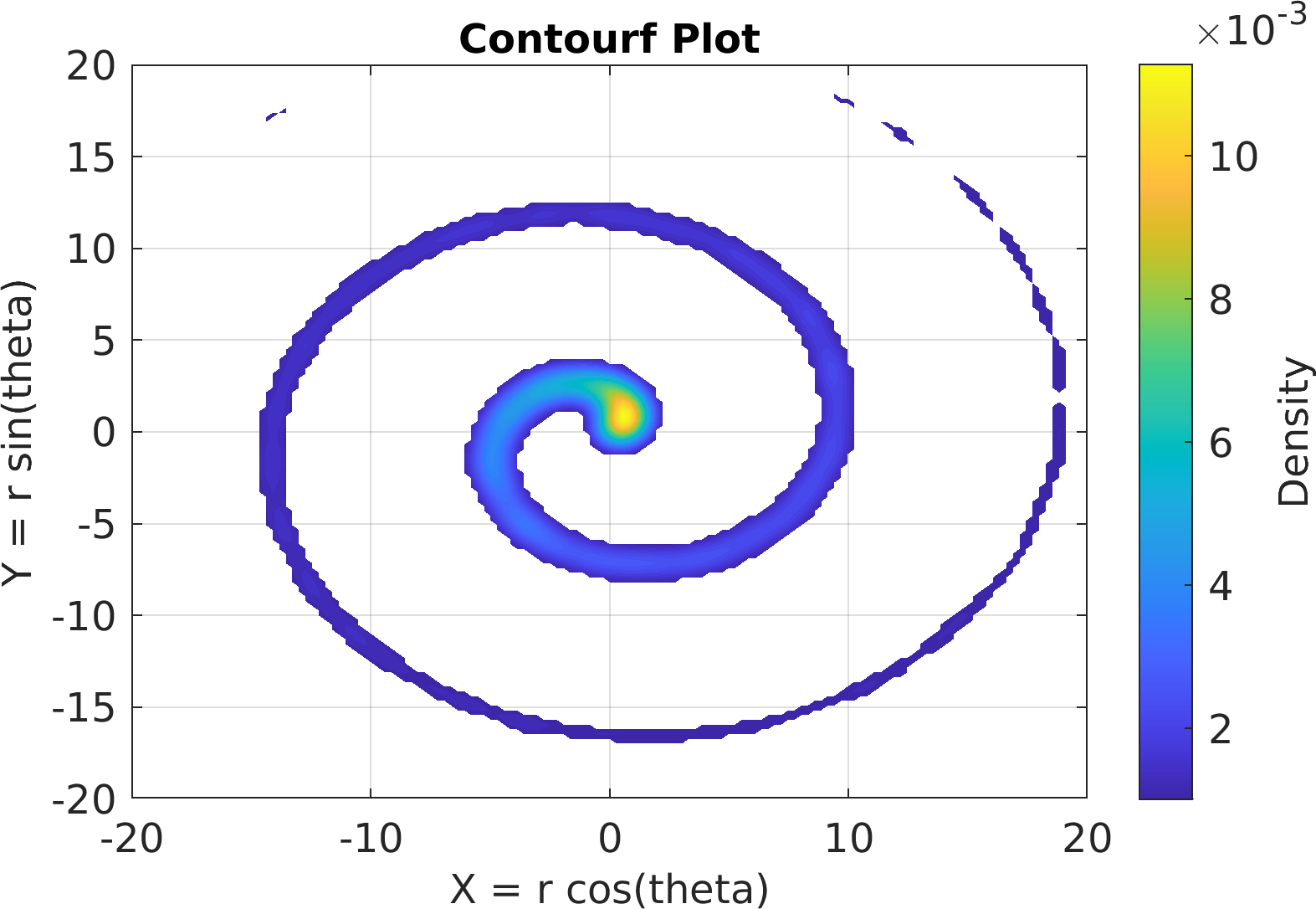
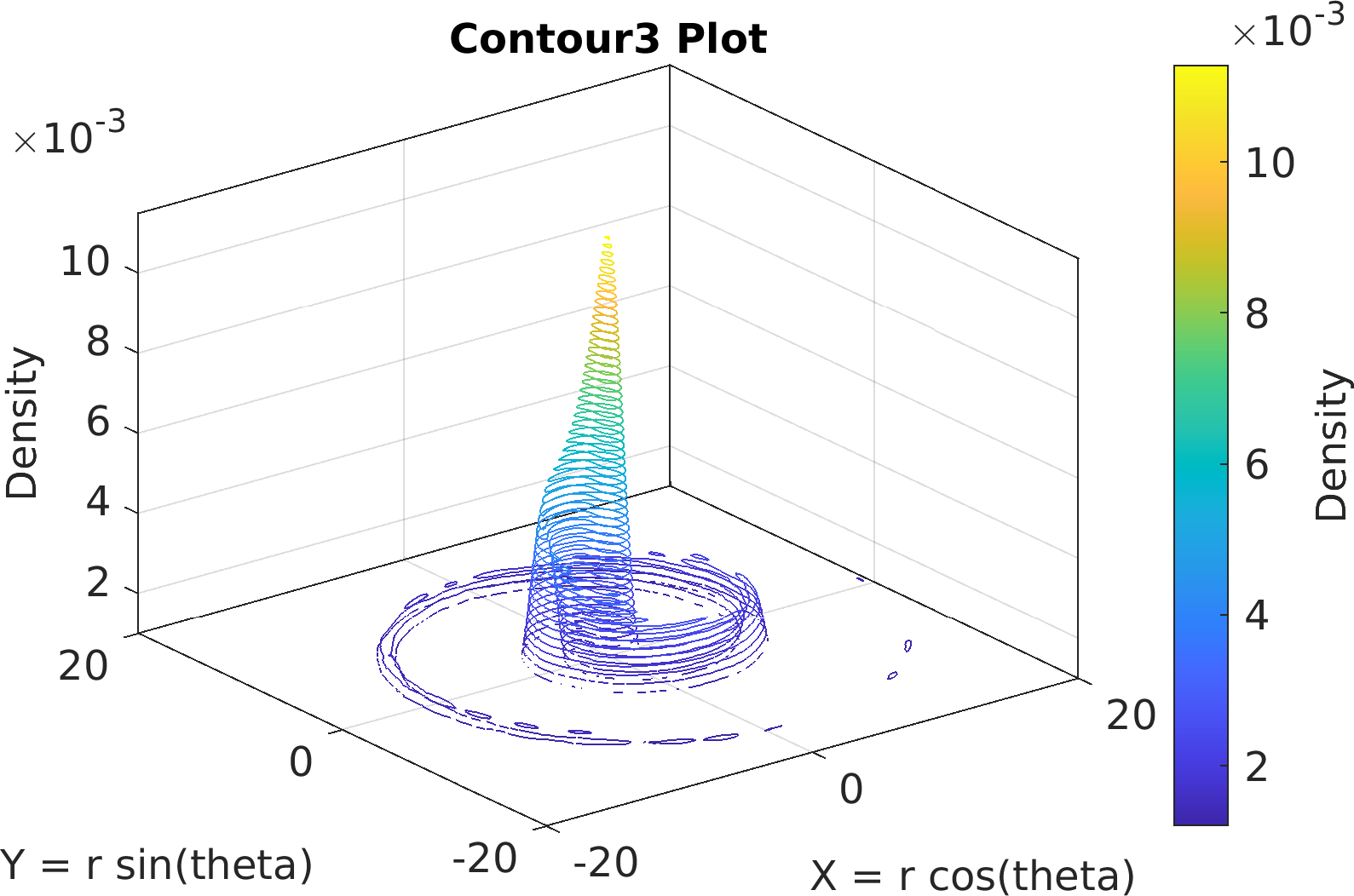
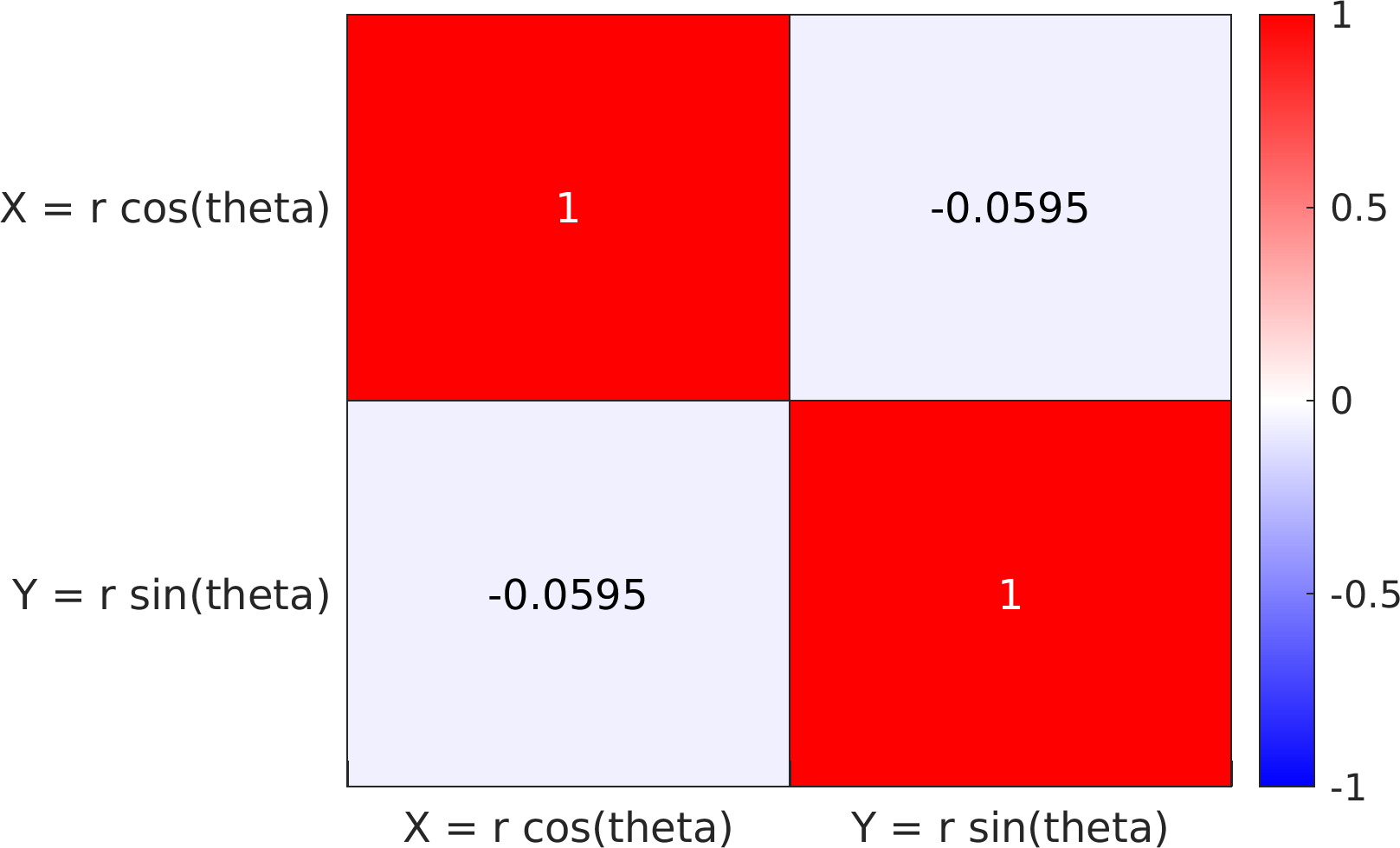
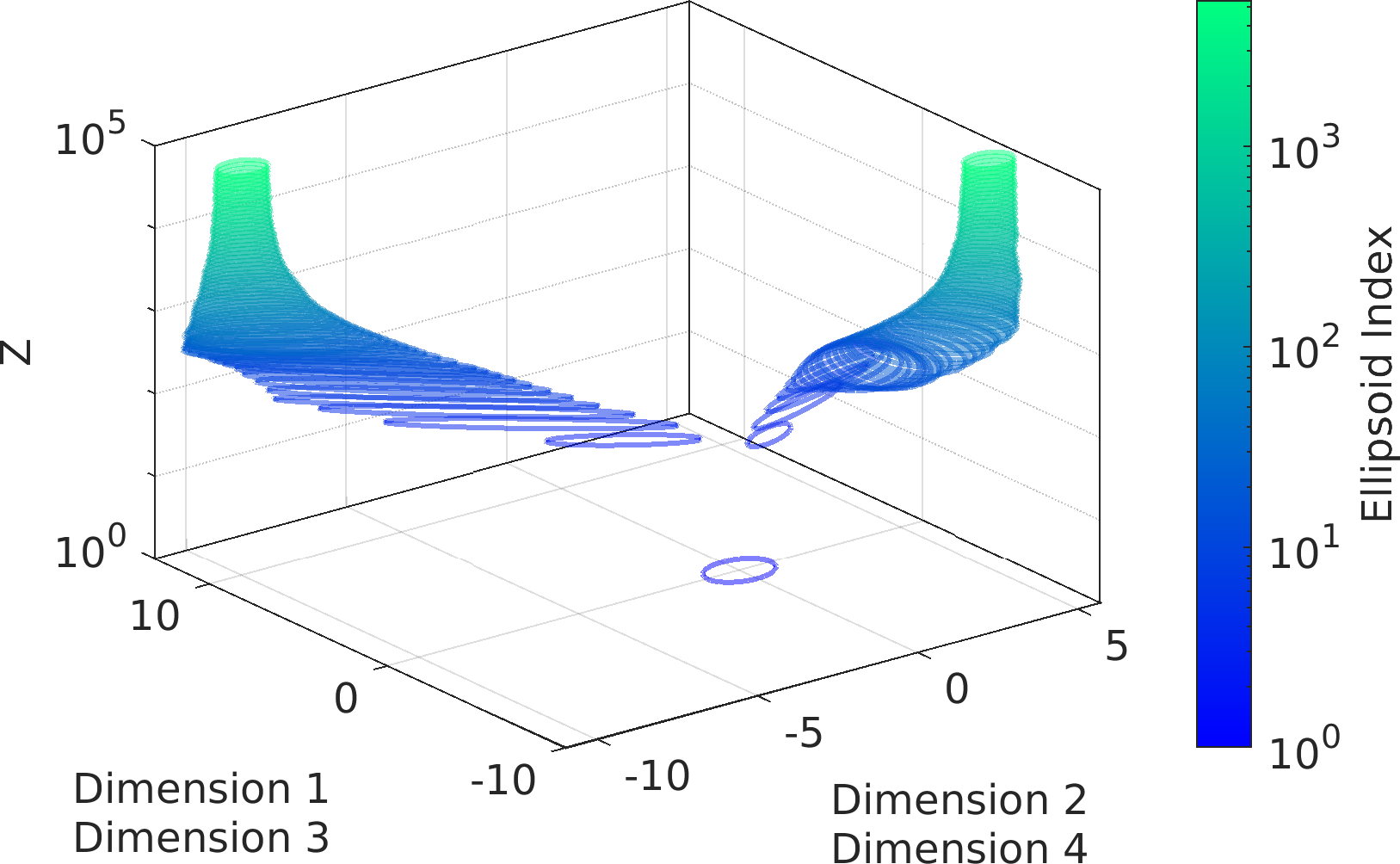
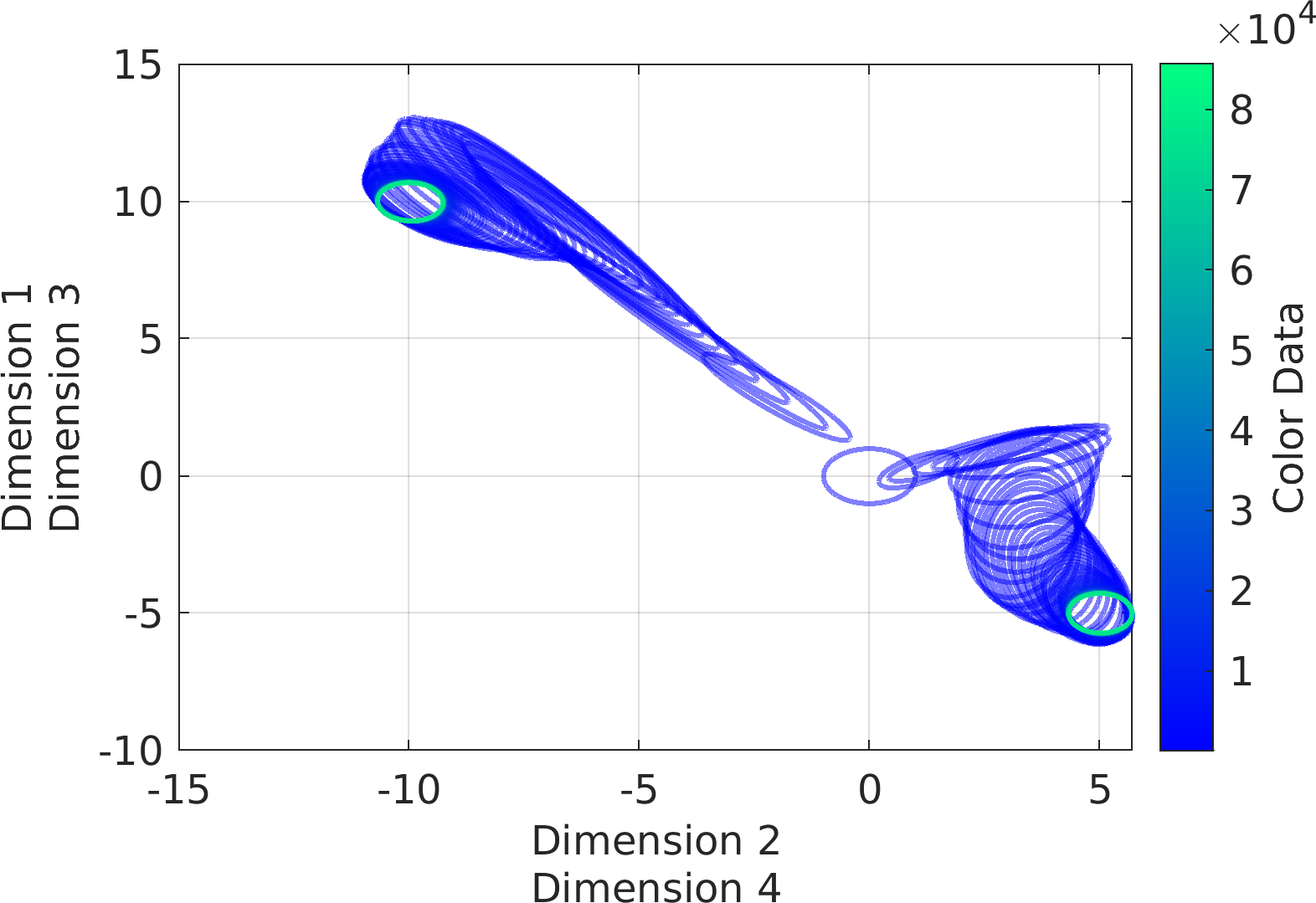
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| function Plot::make | ( | in | self, |
| in | varargin | ||
| ) |
Configure the plot settings and specifications, make the plot, and return nothing.
In making the figure, this method we call the make() methods of each of the subplot objects stored in the subplot component.
| [in,out] | self | : The input/output parent object of class pm.vis.Subplot which is implicitly passed to this dynamic method (not by the user). |
| [in] | varargin | : Any property, value pair of the parent object.If the property is a struct(), then its value must be given as a cell array, with consecutive elements representing the struct property-name, property-value pairs.Note that all of these property-value pairs can be also directly set via the parent object attributes, before calling the make() method. |
Possible calling interfaces ⛓
varargin can also contain the components of the subplot component of the parent object.
Example usage ⛓















Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| function Plot::premake | ( | in | self, |
| in | varargin | ||
| ) |
Configure the plot settings and specifications and return nothing.
| [in,out] | self | : The input/output parent object of class pm.vis.Subplot which is implicitly passed to this dynamic method (not by the user). |
| [in] | varargin | : Any property, value pair of the parent object.If the property is a struct(), then its value must be given as a cell array, with consecutive elements representing the struct property-name, property-value pairs.Note that all of these property-value pairs can be also directly set via the parent object attributes, before calling the premake() method. |
Possible calling interfaces ⛓
Example usage ⛓
f = pm.vis.Plot(pm.vis.Line());
f.premake("figure", {"color", "none"})
| function Plot::reset | ( | in | self, |
| in | varargin | ||
| ) |
Reset the properties of the plot to the original default settings.
Use this method when you change many attributes of the plot and you want to clean up and go back to the default settings.
| [in,out] | self | : The input/output parent object of class pm.vis.Subplot which is implicitly passed to this dynamic method (not by the user). |
| [in] | varargin | : Any property, value pair of the parent object.If the property is a struct(), then its value must be given as a cell array, with consecutive elements representing the struct property-name, property-value pairs.Note that all of these property-value pairs can be also directly set via the parent object attributes, before calling the make() method. |
Possible calling interfaces ⛓
varargin can also contain the components of the subplot component of the parent object.
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| Property Plot::subplot |
subplot
The scalar object of superclass pm.vis.Subplot representing the set of subplots to display in the figure.