This module contains classes and procedures for computing various statistical quantities related to the Exponential distribution.
More...
This module contains classes and procedures for computing various statistical quantities related to the Exponential distribution.
Specifically, this module contains routines for computing the following quantities of the Exponential distribution:
-
the Probability Density Function (PDF)
-
the Cumulative Distribution Function (CDF)
-
the Random Number Generation from the distribution (RNG)
-
the Inverse Cumulative Distribution Function (ICDF) or the Quantile Function
The PDF of the Exponential distribution with the two location and scale parameters \((\mu, \sigma)\) is defined as,
\begin{equation}
\large
F(x | \mu, \sigma) =
\begin{cases}
\frac{1}{\sigma} \exp(-\frac{x - \mu}{\sigma}) &,~ \mu \leq x < +\infty \\
0 &,~ x < \mu
\end{cases}
\end{equation}
where \(-\infty < \mu < +\infty\) is the location parameter and \(0 < \sigma < +\infty\) is the scale parameter of the distribution.
The corresponding CDF with the two location and scale parameters \((\mu, \sigma)\) is defined as,
\begin{equation}
\large
\mathrm{CDF}(x | \mu, \sigma) =
\begin{cases}
1 - \exp(-\frac{x - \mu}{\sigma}) &,~ \mu \leq x < +\infty \\
0 &,~ x < \mu
\end{cases}
\end{equation}
where \(-\infty < \mu < +\infty\) is the location parameter and \(0 < \sigma < +\infty\) is the scale parameter of the distribution.
Random Number Generation
Assuming \(U \in (0, 1]\) is a uniformly-distributed random variate,
\begin{equation}
\large
T = -\sigma\log(U) + \mu ~,
\end{equation}
is a random variate from the Exponential distribution with the location parameter \(-\infty < \mu < +\infty\) and the scale parameter \(0 < \sigma < +\infty\) such that \(\mu \leq T < +\infty\).
- Note
- Note that the mean of the Exponential distribution is the scale parameter: \(\mathrm{mean} = \sigma\)).
- See also
- pm_distUnif
pm_distNegExp
- Benchmarks:
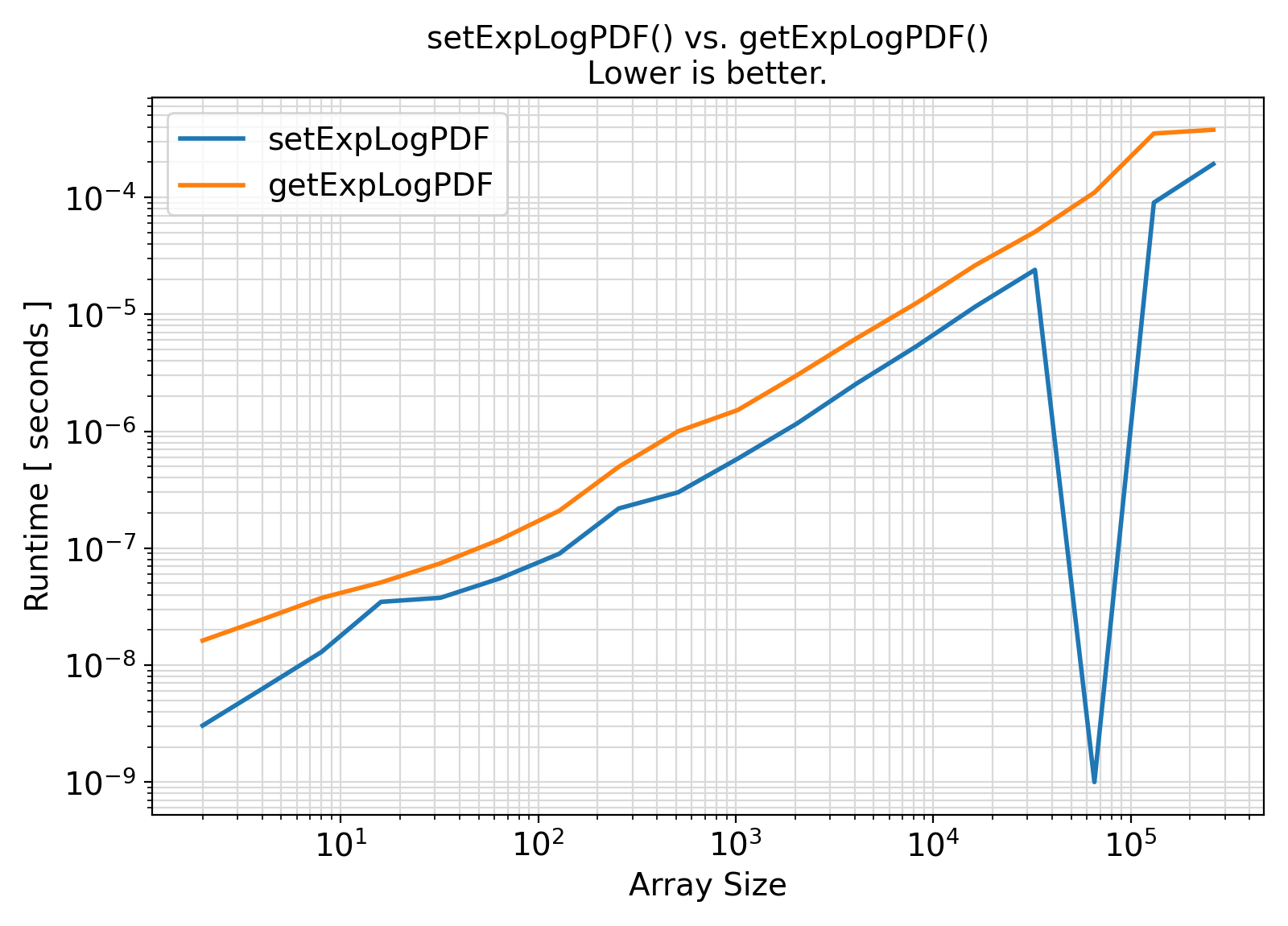
Benchmark :: The runtime performance of getExpLogPDF vs. setExpLogPDF ⛓
4 use iso_fortran_env,
only:
error_unit
12 integer(IK) :: fileUnit
13 integer(IK) ,
parameter :: NSIZE
= 18_IK
14 integer(IK) ,
parameter :: NBENCH
= 2_IK
15 integer(IK) :: arraySize(NSIZE)
16 real(RK) ,
allocatable :: Array(:), Point(:)
17 real(RK) :: dummy
= 0._RK
18 type(bench_type) :: bench(NBENCH)
20 bench(
1)
= bench_type(name
= SK_
"getExpLogPDF", exec
= getExpLogPDF , overhead
= setOverhead)
21 bench(
2)
= bench_type(name
= SK_
"setExpLogPDF", exec
= setExpLogPDF , overhead
= setOverhead)
23 arraySize
= [(
2_IK**isize, isize
= 1_IK, NSIZE )]
25 write(
*,
"(*(g0,:,' '))")
26 write(
*,
"(*(g0,:,' vs. '))") (bench(i)
%name, i
= 1, NBENCH)
27 write(
*,
"(*(g0,:,' '))")
29 open(newunit
= fileUnit, file
= "main.out", status
= "replace")
31 write(fileUnit,
"(*(g0,:,','))")
"arraySize", (bench(i)
%name, i
= 1, NBENCH)
33 loopOverArraySize:
do isize
= 1, NSIZE
35 write(
*,
"(*(g0,:,' '))")
"Benchmarking with size", arraySize(isize)
37 allocate(Array(arraySize(isize)), Point(arraySize(isize)))
39 bench(i)
%timing
= bench(i)
%getTiming(minsec
= 0.05_RK)
41 deallocate(Array, Point)
43 write(fileUnit,
"(*(g0,:,','))") arraySize(isize), (bench(i)
%timing
%mean, i
= 1, NBENCH)
45 end do loopOverArraySize
47 write(
*,
"(*(g0,:,' '))") dummy
48 write(
*,
"(*(g0,:,' '))")
58 subroutine setOverhead()
63 subroutine initialize()
64 call random_number(Point)
68 dummy
= dummy
+ Array(
1)
71 subroutine setExpLogPDF()
80 subroutine getExpLogPDF()
Generate and return an object of type timing_type containing the benchmark timing information and sta...
Generate and return the natural logarithm of the Probability Density Function (PDF) of the Exponentia...
Return the natural logarithm of the Probability Density Function (PDF) of the Exponential distributio...
This module contains abstract interfaces and types that facilitate benchmarking of different procedur...
This module contains classes and procedures for computing various statistical quantities related to t...
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter RK
The default real kind in the ParaMonte library: real64 in Fortran, c_double in C-Fortran Interoperati...
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
This is the class for creating benchmark and performance-profiling objects.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Postprocessing of the benchmark output ⛓
3import matplotlib.pyplot
as plt
9methods = [
"setExpLogPDF",
"getExpLogPDF"]
11df = pd.read_csv(
"main.out")
17ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
21 plt.plot( df[
"arraySize"].values
26plt.xticks(fontsize = fontsize)
27plt.yticks(fontsize = fontsize)
28ax.set_xlabel(
"Array Size", fontsize = fontsize)
29ax.set_ylabel(
"Runtime [ seconds ]", fontsize = fontsize)
30ax.set_title(
"setExpLogPDF() vs. getExpLogPDF()\nLower is better.", fontsize = fontsize)
34plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
35ax.tick_params(axis =
"y", which =
"minor")
36ax.tick_params(axis =
"x", which =
"minor")
44plt.savefig(
"benchmark.getExpLogPDF_vs_setExpLogPDF.runtime.png")
50ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
53plt.plot( df[
"arraySize"].values
54 , np.ones(len(df[
"arraySize"].values))
59plt.plot( df[
"arraySize"].values
60 , df[
"getExpLogPDF"].values / df[
"setExpLogPDF"].values
64plt.xticks(fontsize = fontsize)
65plt.yticks(fontsize = fontsize)
66ax.set_xlabel(
"Array Size", fontsize = fontsize)
67ax.set_ylabel(
"Runtime compared to setExpLogPDF()", fontsize = fontsize)
68ax.set_title(
"getExpLogPDF() / setExpLogPDF()\nLower means faster. Lower than 1 means faster than setExpLogPDF().", fontsize = fontsize)
72plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
73ax.tick_params(axis =
"y", which =
"minor")
74ax.tick_params(axis =
"x", which =
"minor")
75ax.legend ( [
"setExpLogPDF",
"getExpLogPDF"]
82plt.savefig(
"benchmark.getExpLogPDF_vs_setExpLogPDF.runtime.ratio.png")
Visualization of the benchmark output ⛓
Benchmark moral ⛓
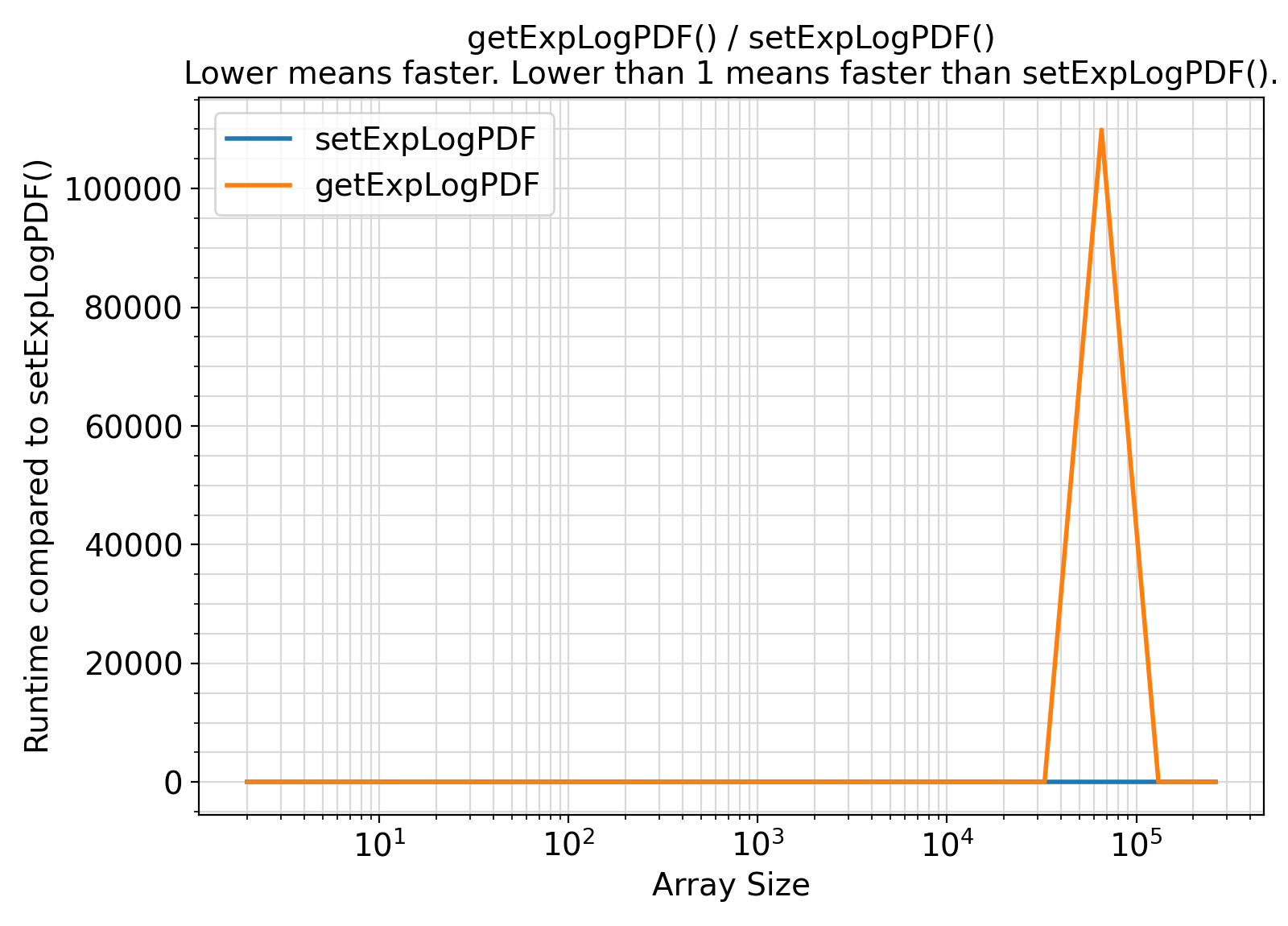
- The procedures under the generic interface getExpLogPDF are functions while the procedures under the generic interface setExpLogPDF are subroutines.
From the benchmark results, it appears that the functional interface performs slightly less efficiently than the subroutine interface when the input array size is small.
Otherwise, the difference appears to be marginal and insignificant in most practical situations.
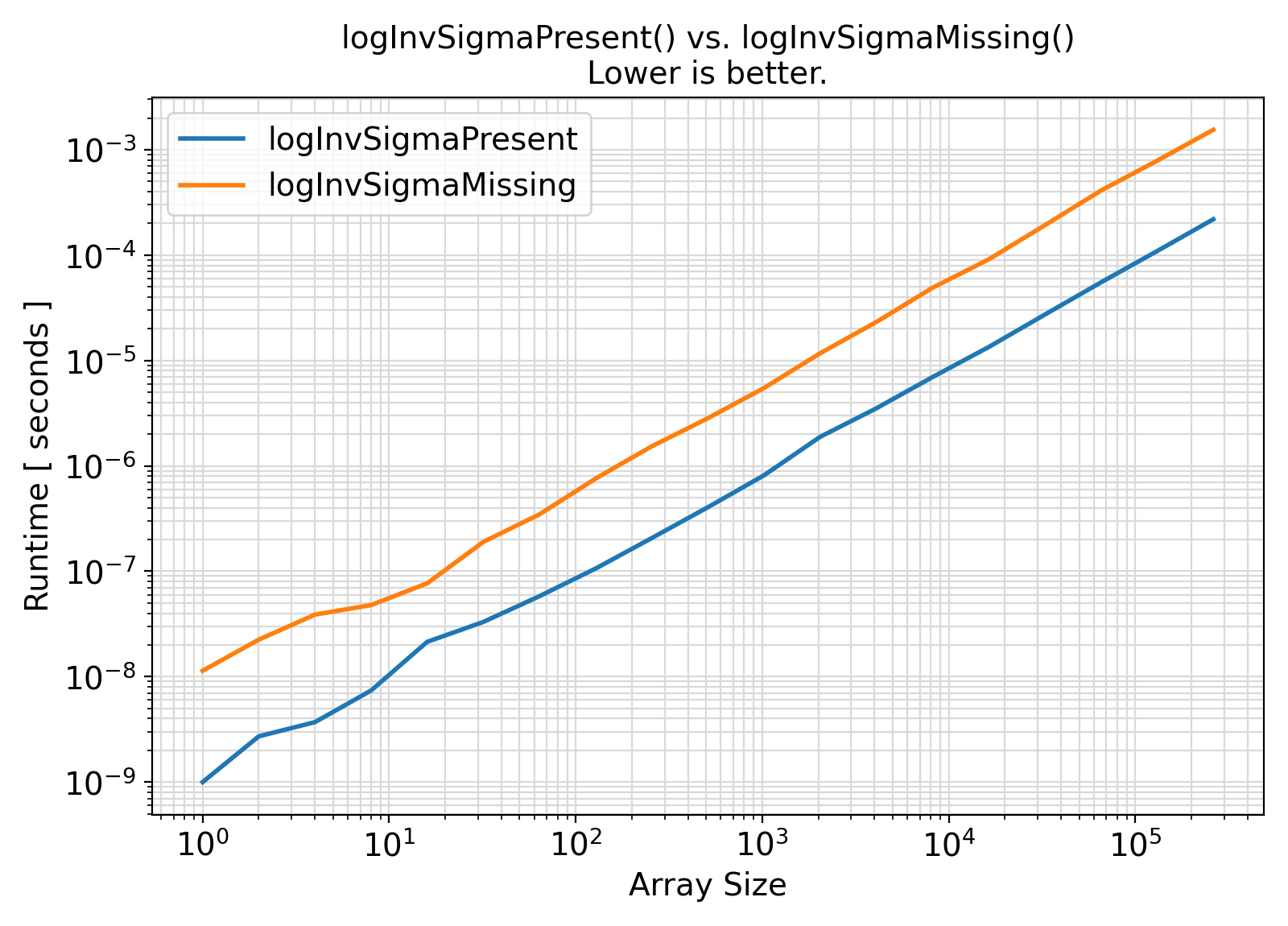
Benchmark :: The runtime performance of setExpLogPDF with and without logInvSigma ⛓
3 use iso_fortran_env,
only:
error_unit
11 integer(IK) :: fileUnit
12 integer(IK) ,
parameter :: NSIZE
= 18_IK
13 integer(IK) ,
parameter :: NBENCH
= 2_IK
14 integer(IK) :: arraySize(
0:NSIZE)
15 real(RK) ,
allocatable :: logPDF(:), point(:)
16 real(RK) ,
allocatable :: logInvSigma(:), invSigma(:)
17 real(RK) :: dummy
= 0._RK
18 type(bench_type) :: bench(NBENCH)
20 bench(
1)
= bench_type(name
= SK_
"logInvSigmaMissing", exec
= logInvSigmaMissing , overhead
= setOverhead)
21 bench(
2)
= bench_type(name
= SK_
"logInvSigmaPresent", exec
= logInvSigmaPresent , overhead
= setOverhead)
23 arraySize
= [(
2_IK**isize, isize
= 0_IK, NSIZE )]
25 write(
*,
"(*(g0,:,' '))")
26 write(
*,
"(*(g0,:,' vs. '))") (bench(i)
%name, i
= 1, NBENCH)
27 write(
*,
"(*(g0,:,' '))")
29 open(newunit
= fileUnit, file
= "main.out", status
= "replace")
31 write(fileUnit,
"(*(g0,:,','))")
"arraySize", (bench(i)
%name, i
= 1, NBENCH)
33 loopOverArraySize:
do isize
= 0, NSIZE
35 write(
*,
"(*(g0,:,' '))")
"Benchmarking with size", arraySize(isize)
37 allocate(logPDF(arraySize(isize)), point(arraySize(isize)), invSigma(arraySize(isize)))
38 call random_number(point)
39 call random_number(invSigma)
40 logInvSigma
= log(invSigma)
42 bench(i)
%timing
= bench(i)
%getTiming(minsec
= 0.05_RK)
44 deallocate(logPDF, point, invSigma)
46 write(fileUnit,
"(*(g0,:,','))") arraySize(isize), (bench(i)
%timing
%mean, i
= 1, NBENCH)
48 end do loopOverArraySize
50 write(
*,
"(*(g0,:,' '))") dummy
51 write(
*,
"(*(g0,:,' '))")
61 subroutine setOverhead()
66 subroutine initialize()
72 dummy
= dummy
+ logPDF(
1)
75 subroutine logInvSigmaPresent()
78 if (arraySize(isize)
> 1_IK)
then
81 call setExpLogPDF(logPDF(
1), point(
1), invSigma(
1), logInvSigma(
1))
86 subroutine logInvSigmaMissing()
89 if (arraySize(isize)
> 1_IK)
then
92 logPDF(
1)
= getExpLogPDF(point(
1), invSigma
= invSigma(
1))
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Postprocessing of the benchmark output ⛓
3import matplotlib.pyplot
as plt
9methods = [
"logInvSigmaPresent",
"logInvSigmaMissing"]
11df = pd.read_csv(
"main.out")
17ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
21 plt.plot( df[
"arraySize"].values
26plt.xticks(fontsize = fontsize)
27plt.yticks(fontsize = fontsize)
28ax.set_xlabel(
"Array Size", fontsize = fontsize)
29ax.set_ylabel(
"Runtime [ seconds ]", fontsize = fontsize)
30ax.set_title(
"logInvSigmaPresent() vs. logInvSigmaMissing()\nLower is better.", fontsize = fontsize)
34plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
35ax.tick_params(axis =
"y", which =
"minor")
36ax.tick_params(axis =
"x", which =
"minor")
44plt.savefig(
"benchmark.setExpLogPDF-logInvSigma-missing_vs_present.runtime.png")
50ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
53plt.plot( df[
"arraySize"].values
54 , np.ones(len(df[
"arraySize"].values))
59plt.plot( df[
"arraySize"].values
60 , df[
"logInvSigmaMissing"].values / df[
"logInvSigmaPresent"].values
64plt.xticks(fontsize = fontsize)
65plt.yticks(fontsize = fontsize)
66ax.set_xlabel(
"Array Size", fontsize = fontsize)
67ax.set_ylabel(
"Runtime compared to logInvSigmaPresent()", fontsize = fontsize)
68ax.set_title(
"logInvSigmaMissing / logInvSigmaPresent\nLower means faster. Lower than 1 means faster than logInvSigmaPresent.", fontsize = fontsize)
72plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
73ax.tick_params(axis =
"y", which =
"minor")
74ax.tick_params(axis =
"x", which =
"minor")
75ax.legend ( [
"logInvSigmaPresent",
"logInvSigmaMissing"]
82plt.savefig(
"benchmark.setExpLogPDF-logInvSigma-missing_vs_present.runtime.ratio.png")
Visualization of the benchmark output ⛓
Benchmark moral ⛓
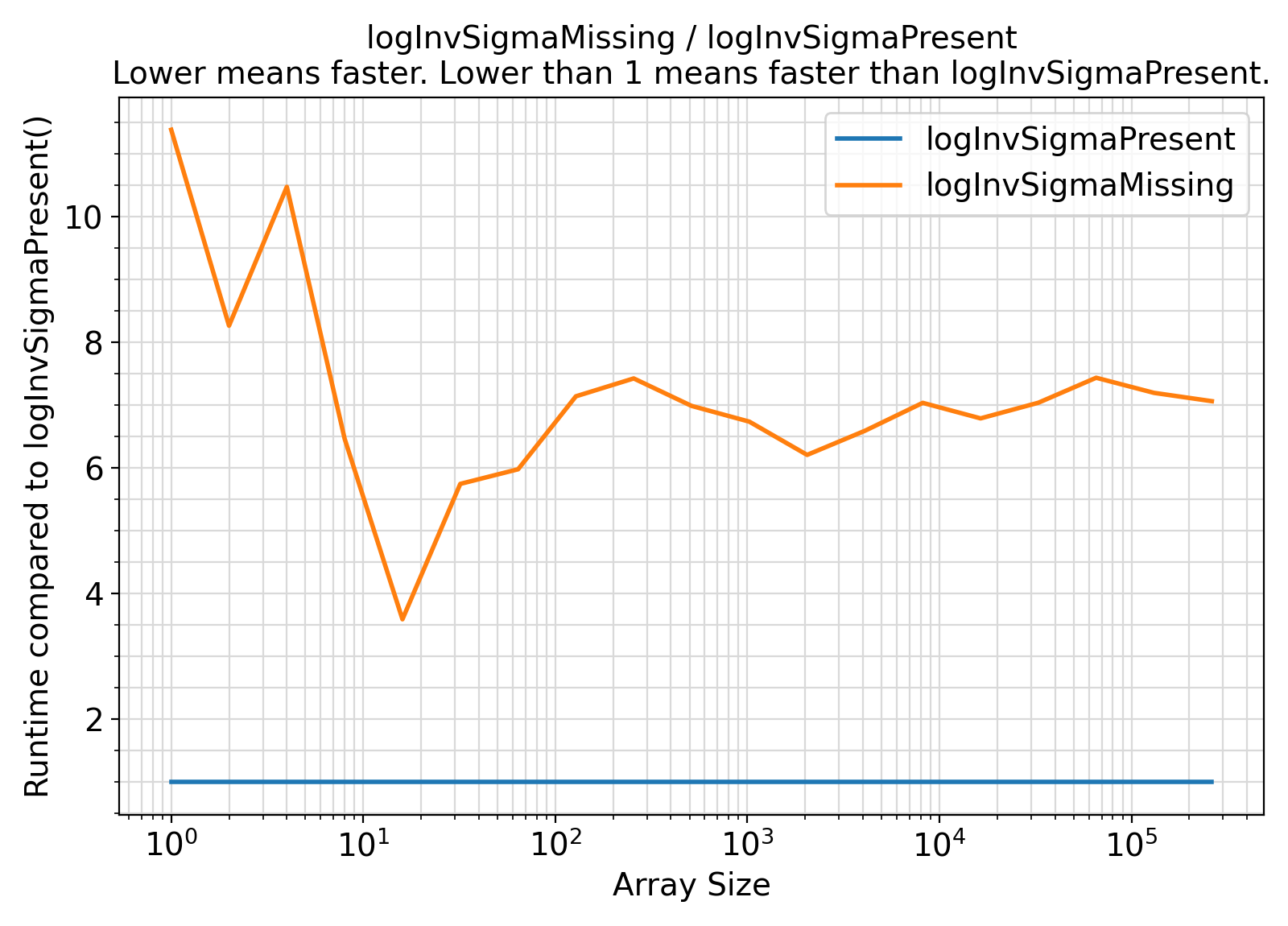
- The procedures under the generic interface setExpLogPDF accept an extra argument
logInvSigma = log(invSigma) while the procedures under the generic interface getExpLogPDF compute this term internally with every procedure call.
In the presence of this argument, the logarithmic computation log(invSigma) will be avoided.
As such, the presence of logInvSigma is expected to lead to faster computations.
- Test:
- test_pm_distExp
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, Oct 16, 2009, 11:14 AM, Michigan