
|
ParaMonte Fortran 2.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |

|
ParaMonte Fortran 2.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
This module contains classes and procedures for computing the properties related to the covariance matrices of a random sample. More...
Data Types | |
| interface | getCov |
Generate and return the (optionally unbiased) covariance matrix of a pair of (potentially weighted) time series x(1:nsam) and y(1:nsam) or of an input (potentially weighted) array of shape (ndim, nsam) or (nsam, ndim) where ndim is the number of data dimensions (the number of data attributes) and nsam is the number of data points.More... | |
| interface | getCovMerged |
| Generate and return the merged covariance of a sample resulting from the merger of two separate (potentially weighted) samples \(A\) and \(B\). More... | |
| interface | setCov |
Return the covariance matrix corresponding to the input (potentially weighted) correlation matrix or return the biased sample covariance matrix of the input array of shape (ndim, nsam) or (nsam, ndim) or a pair of (potentially weighted) time series x(1:nsam) and y(1:nsam) where ndim is the number of data dimensions (the number of data attributes) and nsam is the number of data points.More... | |
| interface | setCovMean |
Return the covariance matrix and mean vector corresponding to the input (potentially weighted) input sample of shape (ndim, nsam) or (nsam, ndim) or a pair of (potentially weighted) time series x(1:nsam) and y(1:nsam) where ndim is the number of data dimensions (the number of data attributes) and nsam is the number of data points.More... | |
| interface | setCovMeanMerged |
| Return the merged covariance and mean of a sample resulting from the merger of two separate (potentially weighted) samples \(A\) and \(B\). More... | |
| interface | setCovMeanUpdated |
| Return the covariance and mean of a sample that results from the merger of two separate (potentially weighted) non-singular \(A\) and singular \(B\) samples. More... | |
| interface | setCovMerged |
| Return the merged covariance of a sample resulting from the merger of two separate (potentially weighted) samples \(A\) and \(B\). More... | |
| interface | setCovUpdated |
| Return the covAariance resulting from the merger of two separate (potentially weighted) non-singular and singular samples \(A\) and \(B\). More... | |
Variables | |
| character(*, SK), parameter | MODULE_NAME = "@pm_sampleCov" |
This module contains classes and procedures for computing the properties related to the covariance matrices of a random sample.
The concept of variance can be generalized to measure the covariation of any pair of data attributes.
The sample covariance matrix is a \(K\)-by- \(K\) matrix \(\mathbf{Q} = \left[\tilde\Sigma_{jk}\right]\) with entries,
\begin{equation} \tilde\Sigma_{jk} = \frac{1}{n} \sum_{i=1}^{n} \left( x_{ij} - \hat\mu_j \right) \left( x_{ik} - \hat\mu_k \right) ~, \end{equation}
where \(n\) is the number of observations in the sample, \(\hat\mu\) is the sample mean vector, and \(\Sigma_{jk}\) is an estimate of the covariance between the \(j\)th variable and the kth variable of the population underlying the data.
The diagonal elements of the matrix \(\tilde\Sigma_{jj}\) are known as the sample variance.
The above formula yields a biased estimate of the covariance matrix of the sample.
Intuitively, the sample covariance relies on the difference between each observation and the sample mean, but the sample mean is slightly correlated with each observation since it is defined in terms of all observations.
Therefore, unless the sample mean is known a priori, the above equation yields a biased estimate of the covariance with sample mean as a proxy for the true mean of the population.
Note that the bias is noticeable only when the sample size is small (e.g., \(<10\)).
A popular fix to the definition of sample covariance to remove its bias is to apply the Bessel correction to the equation above, yielding the unbiased covariance estimate as,
\begin{eqnarray} \hat\Sigma_{jk} &=& \frac{\xi}{n} \sum_{i=1}^{n} \left( x_{ij} - \hat\mu_j \right) \left( x_{ik} - \hat\mu_k \right) ~, &=& \frac{1}{n - 1} \sum_{i=1}^{n} \left( x_{ij} - \hat\mu_j \right) \left( x_{ik} - \hat\mu_k \right) ~, \end{eqnarray}
where \(\xi = \frac{n}{n - 1}\) is the Bessel bias correction factor.
\begin{equation} \tilde{\Sigma}^w_{jk} = \frac{ \sum_{i = 1}^{n} \left( x_{ij} - \hat\mu^w_j \right) \left( x_{ik} - \hat\mu^w_k \right) } { \left( \sum_{i=1}^{n} w_i \right) } ~. \end{equation}
where n = nsam is the number of observations in the sample, \(w_i\) are the weights of individual data points, the superscript \(^w\) signifies the sample weights, and \(\hat\mu^w\) is the weighted mean of the sample.
When the sample size is small, the above equation yields a biased estimate of the covariance.
Unbiased weighted sample covariance
There is no unique generic equation for the unbiased covariance of a weighted sample.
However, depending on the types of the weights involved, a few popular definitions exist.
\begin{equation} \hat\Sigma^w_{jk} = \frac{ \sum_{i = 1}^{n} \left( x_{ij} - \hat\mu^w_j \right) \left( x_{ik} - \hat\mu^w_k \right) } { \left( \sum_{i=1}^{n} w_i \right) - 1} ~. \end{equation}
Frequency weights represent the number of duplications of each observation in the sample whose population covariance is to be estimated.\begin{equation} \hat\Sigma^w_{jk} = \frac{ \sum_{i=1}^{n} w_i } { \left( \sum_{i=1}^{n} w_i \right)^2 - \left( \sum_{i=1}^{n} w_i^2 \right) } \sum_{i = 1}^{n} \left( x_{ij} - \hat\mu^w_j \right) \left( x_{ik} - \hat\mu^w_k \right) ~. \end{equation}
The covariance matrix \(\Sigma\) is related to the correlation matrix \(\rho\) by the following equation,
\begin{equation} \Sigma_{ij} = \rho_{ij} \times \sigma_{i} \times \sigma_{j} ~, \end{equation}
where \(\Sigma\) represents the covariance matrix, \(\rho\) represents the correlation matrix, and \(\sigma\) represents the standard deviations.
Benchmark :: The runtime performance of setCov vs. setCovMean. ⛓
ifort compiler ⛓ ifort compiler ⛓ gfortran compiler ⛓ 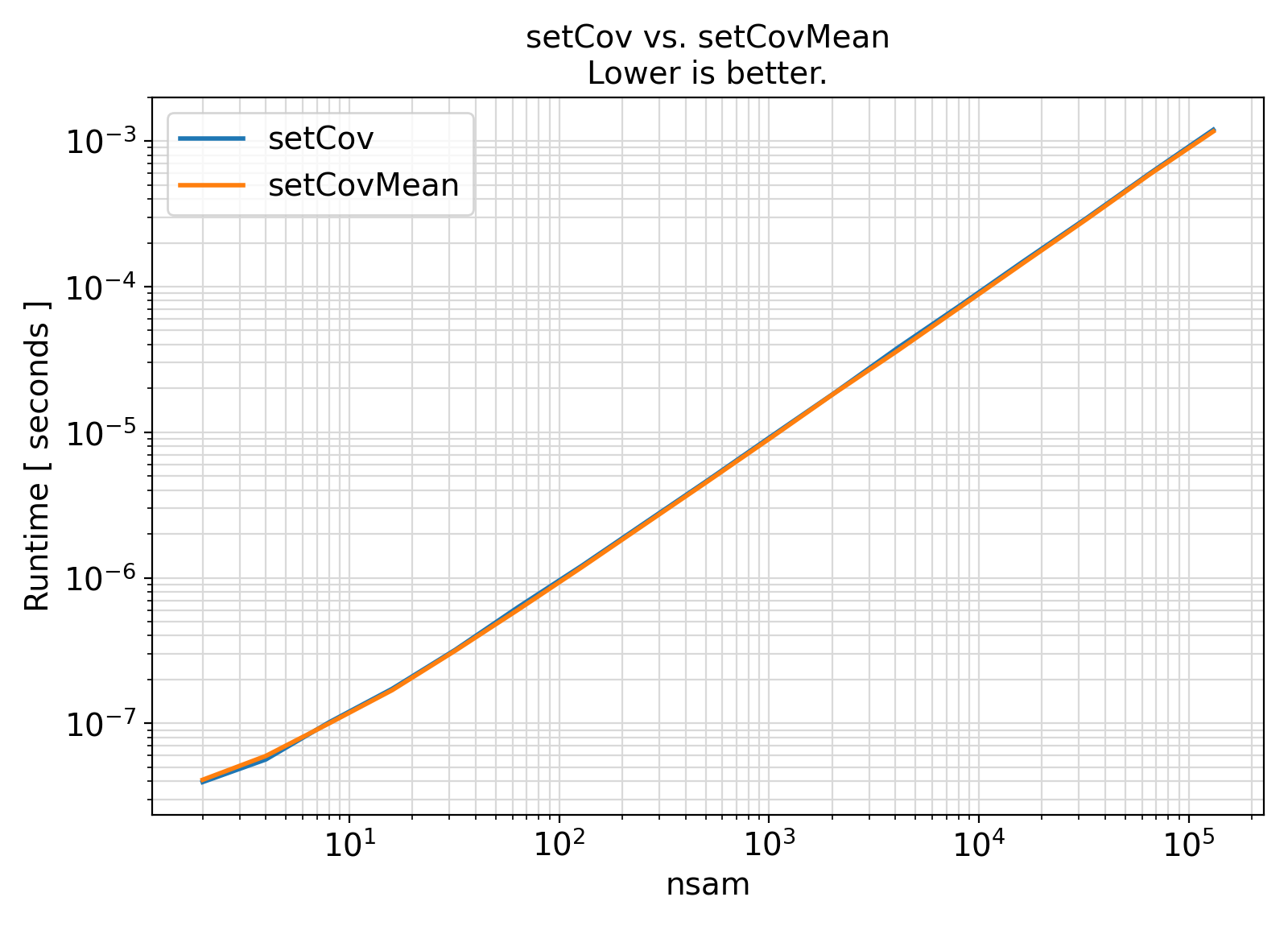
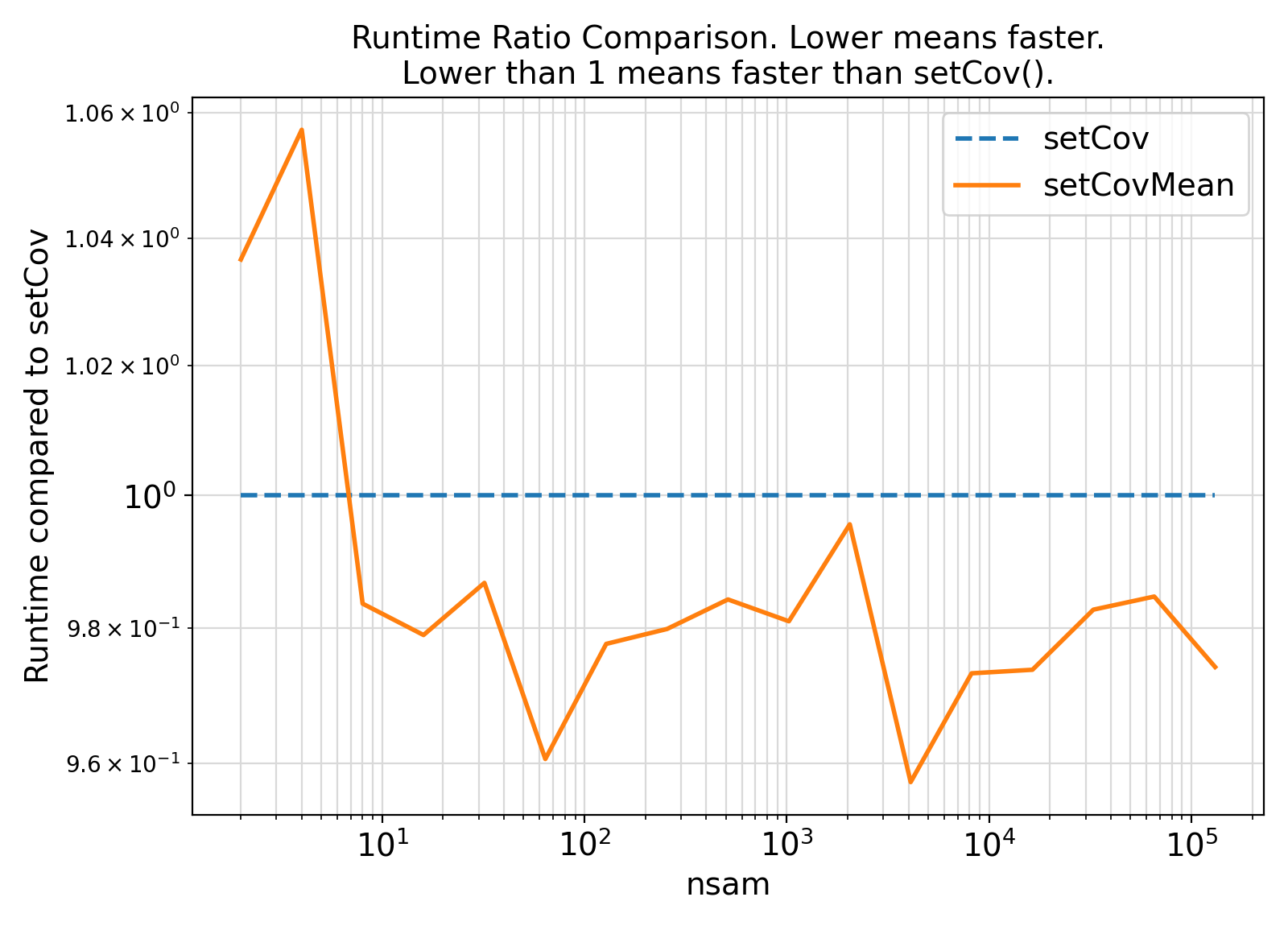
gfortran gfortran.gfortran support, separate generic interfaces were instead developed for different sample weight types.gfortran PDT bugs are resolved, the getVar generic interface can be extended to serve as a high-level wrapper for the weight-specific generic interfaces in this module.
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| character(*, SK), parameter pm_sampleCov::MODULE_NAME = "@pm_sampleCov" |
Definition at line 182 of file pm_sampleCov.F90.