This module contains procedures and generic interfaces for computing the Cholesky factorization of positive definite matrices.
More...
This module contains procedures and generic interfaces for computing the Cholesky factorization of positive definite matrices.
Quick start
The recommended routines for computing the Cholesky factorization are the procedures under the generic interface setChoLow() with assumed-shape dummy arguments.
The explicit-shape procedures are not recommended as the array bounds cannot be checked and the return of error status for the Cholesky factorization failure is implicit (via the first element of the output argument dia).
Cholesky factorization
In linear algebra, the Cholesky decomposition or Cholesky factorization is a decomposition of a Hermitian, positive-definite matrix into the product of a lower triangular matrix and its conjugate transpose.
It was discovered by André-Louis Cholesky for real matrices, and posthumously published in 1924.
Algorithmic efficiency
When it is applicable, the Cholesky decomposition is roughly twice as efficient as the LU decomposition for solving systems of linear equations.
Definition
The Cholesky decomposition of a Hermitian positive-definite matrix A is a decomposition of the form,
\begin{equation}
\mathbf{A} = \mathbf{LL}^{*} ~,
\end{equation}
where L is a lower triangular matrix with real and positive diagonal entries, and L* denotes the conjugate transpose of L.
Every Hermitian positive-definite matrix (and thus also every real-valued symmetric positive-definite matrix) has a unique Cholesky decomposition.
The converse holds trivially: if A can be written as LL* for some invertible L, lower triangular or otherwise, then A is Hermitian and positive definite.
When A is a real matrix (hence symmetric positive-definite), the factorization may be written as,
\begin{equation}
\mathbf{A} = \mathbf{LL}^{\mathsf{T}} ~,
\end{equation}
where L is a real lower triangular matrix with positive diagonal entries.
Algorithm
For complex Hermitian matrix, the following formula applies
\begin{eqnarray}
L_{j,j} &=& \sqrt{A_{j,j} - \sum_{k=1}^{j-1}L_{j,k}^{*}L_{j,k}} ~, \\
L_{i,j} &=& \frac{1}{L_{j,j}} \left(A_{i,j} - \sum_{k=1}^{j-1}L_{j,k}^{*}L_{i,k}\right) \quad {\text{for }} i > j ~.
\end{eqnarray}
Therefore, the computation of the entry (i, j) depends on the entries to the left and above.
The computation is usually arranged in either of the following orders:
-
The Cholesky–Banachiewicz algorithm (row-major) starts from the upper left corner of the matrix L and proceeds to calculate the matrix row by row:
do i = 1, size(A,1)
L(i,i) = sqrt(A(i,i) - dot_product(L(i,1:i-1), L(i,1:i-1)))
L(i+1:,i) = (A(i+1:,i) - matmul(conjg(L(i,1:i-1)), L(i+1:,1:i-1))) / L(i,i)
end do
-
The Cholesky–Crout algorithm starts from the upper left corner of the matrix L and proceeds to calculate the matrix column by column:
do i = 1, size(A,1)
L(i,i) = sqrt(A(i,i) - dot_product(L(1:i-1,i), L(1:i-1,i)))
L(i,i+1:) = (A(i,i+1:) - matmul(conjg(L(1:i-1,i)), L(1:i-1,i+1:))) / L(i,i)
end do
Either pattern of access allows the entire computation to be performed in-place if desired.
Given that Fortran is a column-major language, the Cholesky–Crout algorithm algorithm can potentially be faster on the contemporary hardware.
This means that the computations can be done more efficiently if one uses the upper-diagonal triangle of a Hermitian matrix to compute the corresponding upper-diagonal triangular Cholesky factorization.
- Benchmarks:
Benchmark :: Cholesky factorization - assumed-shape vs. explicit-shape dummy arguments ⛓
- Here is a code snippet to compare the performances of the Cholesky factorization routine setChoLow() via two different interfaces:
-
Assumed-shape dummy arguments with explicit return of failure error flag (recommended).
-
Explicit-shape dummy arguments with implicit return of failure error flag (not recommended).
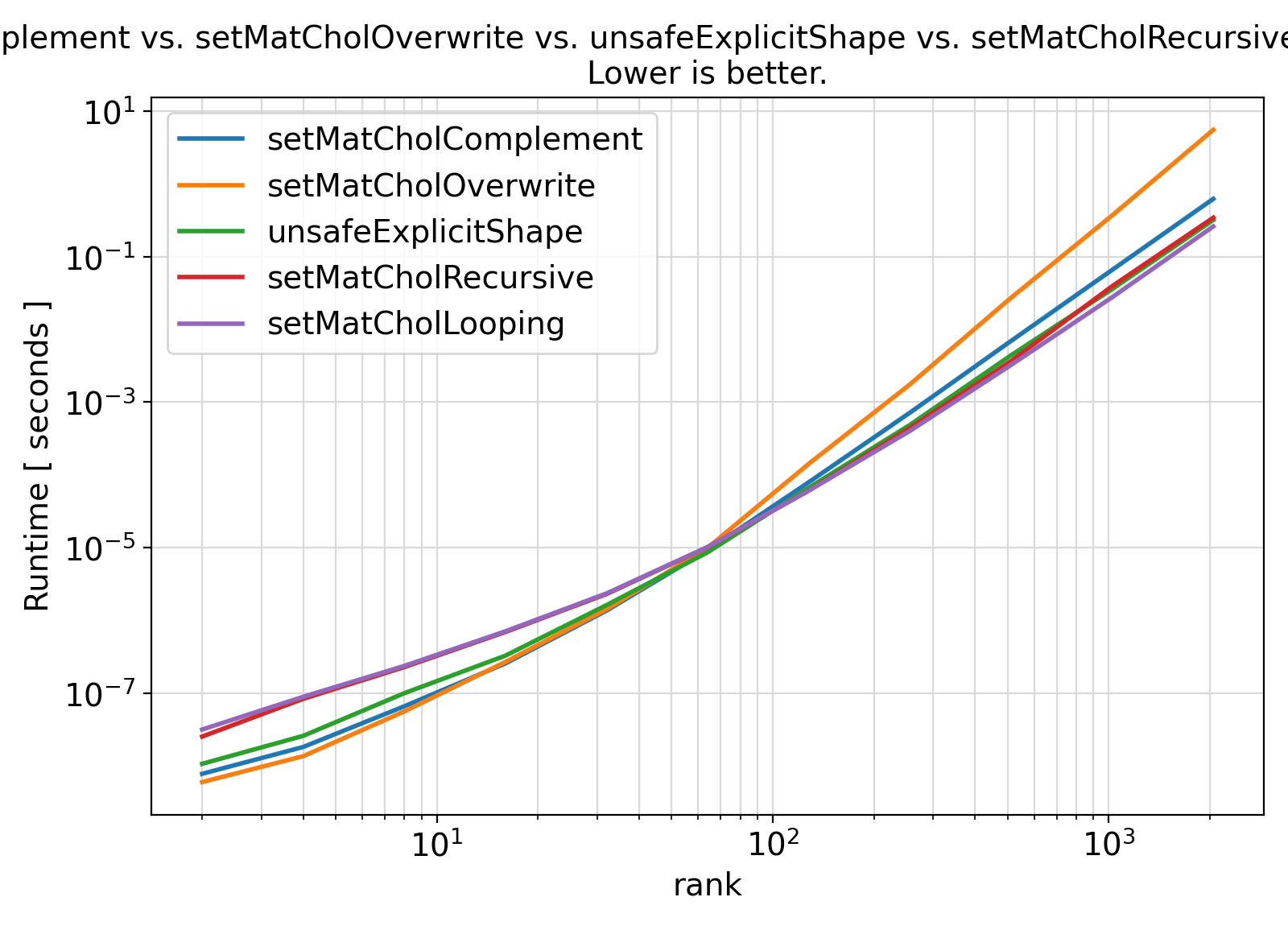
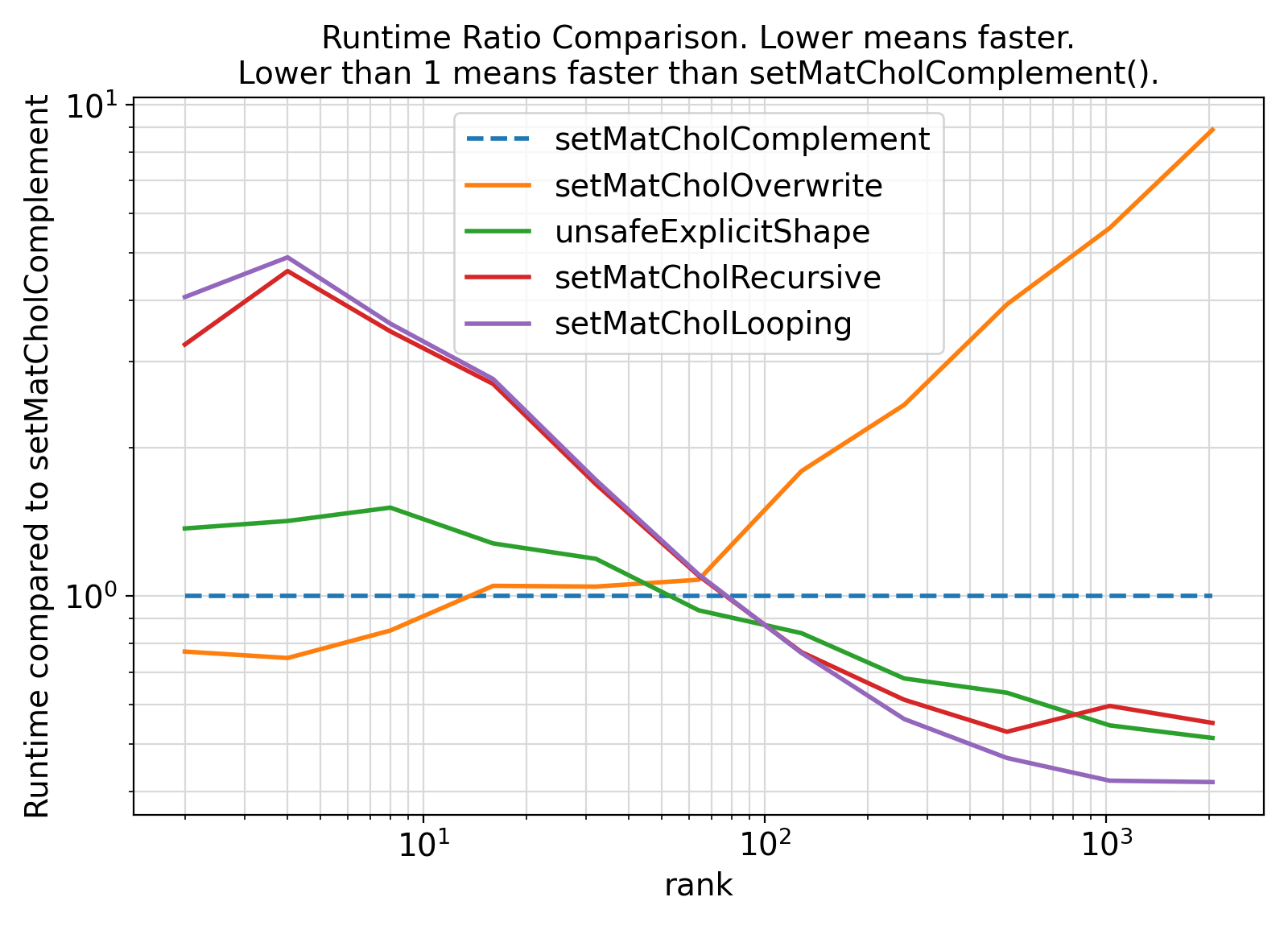
Overall, no significant difference between the two approaches is observed, meaning that the safe interface with assumed-shape arrays is much better to use.
11 integer(IK) :: itry, ntry
14 integer(IK) :: fileUnit
15 integer(IK) ,
parameter :: NARR
= 11_IK
16 real(RKG) ,
allocatable :: mat(:,:), choDia(:)
17 type(bench_type),
allocatable :: bench(:)
18 integer(IK) ,
parameter :: nsim
= 2**NARR
20 type(subset_type),
parameter :: subset
= subset_type()
23 offset
= merge(
1,
0,
same_type_as(subset, lowDia_type()))
25 bench
= [
bench_type(name
= SK_
"setMatCholComplement", exec
= setMatCholComplement, overhead
= setOverhead)
&
26 ,
bench_type(name
= SK_
"setMatCholOverwrite", exec
= setMatCholOverwrite, overhead
= setOverhead)
&
27 ,
bench_type(name
= SK_
"unsafeExplicitShape", exec
= unsafeExplicitShape, overhead
= setOverhead)
&
28 ,
bench_type(name
= SK_
"setMatCholRecursive", exec
= setMatCholRecursive, overhead
= setOverhead)
&
29 ,
bench_type(name
= SK_
"setMatCholLooping", exec
= setMatCholLooping, overhead
= setOverhead)
&
31 ,
bench_type(name
= SK_
"lapack_dpotrf", exec
= lapack_dpotrf, overhead
= setOverhead)
&
35 write(
*,
"(*(g0,:,' '))")
36 write(
*,
"(*(g0,:,' '))")
"assumed-shape vs. explicit-shape setChoLow()."
37 write(
*,
"(*(g0,:,' '))")
39 open(newunit
= fileUnit, file
= "main.out", status
= "replace")
41 write(fileUnit,
"(*(g0,:,','))")
"rank", (bench(i)
%name, i
= 1,
size(bench))
44 loopOverMatrixSize:
do iarr
= 1, NARR
48 allocate(mat(rank,
0 : rank), choDia(rank))
49 write(
*,
"(*(g0,:,' '))")
"Benchmarking setChoLow() algorithms with array size", rank, ntry
55 write(fileUnit,
"(*(g0,:,','))") rank, (bench(i)
%timing
%mean
/ ntry, i
= 1,
size(bench))
56 deallocate(mat, choDia)
58 end do loopOverMatrixSize
59 write(
*,
"(*(g0,:,' '))")
69 subroutine setOverhead()
75 subroutine getMatrix()
77 call random_number(mat)
79 do i
= 1,
size(mat,
dim = 1,
kind = IK)
80 mat(i, i
- offset)
= 1._RKG
86 subroutine lapack_dpotrf()
90 call dpotrf(
"U", rank, mat(
1,
1), rank, info)
91 if (info
/= 0_IK)
error stop
96 subroutine unsafeExplicitShape()
101 call setChoLow(mat(:,
1-offset:rank
-offset), choDia, rank)
102 if (choDia(
1)
< 0._RKG)
error stop
106 subroutine setMatCholOverwrite()
111 call setMatChol(mat(:,
1-offset:rank
-offset), subset, info, mat(:,
1-offset:rank
-offset), nothing)
112 if (info
/= 0_IK)
error stop
116 subroutine setMatCholComplement()
122 call setMatChol(mat(:,
1-offset:rank
-offset), subset, info, mat(:,offset:rank), transHerm)
123 if (info
/= 0_IK)
error stop
127 subroutine setMatCholLooping()
132 call setMatChol(mat(:,
1-offset:rank
-offset), subset, info, iteration)
133 if (info
/= 0_IK)
error stop
137 subroutine setMatCholRecursive()
142 call setMatChol(mat(:,
1-offset:rank
-offset), subset, info, recursion)
143 if (info
/= 0_IK)
error stop
Generate and return an object of type timing_type containing the benchmark timing information and sta...
[LEGACY code] Return the lower-triangle of the Cholesky factorization of the symmetric positive-def...
Compute and return the lower/upper-triangle of the Cholesky factorization of the input Symmetric/Her...
This module contains abstract interfaces and types that facilitate benchmarking of different procedur...
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter RKD
The double precision real kind in Fortran mode. On most platforms, this is an 64-bit real kind.
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
This module contains procedures and generic interfaces for computing the Cholesky factorization of po...
This is the class for creating benchmark and performance-profiling objects.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Postprocessing of the benchmark output ⛓
3import matplotlib.pyplot
as plt
8dirname = os.path.basename(os.getcwd())
12df = pd.read_csv(
"main.out", delimiter =
",")
13colnames = list(df.columns.values)
19ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
22for colname
in colnames[1:]:
23 plt.plot( df[colnames[0]].values
28plt.xticks(fontsize = fontsize)
29plt.yticks(fontsize = fontsize)
30ax.set_xlabel(colnames[0], fontsize = fontsize)
31ax.set_ylabel(
"Runtime [ seconds ]", fontsize = fontsize)
32ax.set_title(
" vs. ".join(colnames[1:])+
"\nLower is better.", fontsize = fontsize)
36plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
37ax.tick_params(axis =
"y", which =
"minor")
38ax.tick_params(axis =
"x", which =
"minor")
39ax.legend ( colnames[1:]
46plt.savefig(
"benchmark." + dirname +
".runtime.png")
52ax = plt.figure(figsize = 1.25 * np.array([6.4,4.6]), dpi = 200)
55plt.plot( df[colnames[0]].values
56 , np.ones(len(df[colnames[0]].values))
61for colname
in colnames[2:]:
62 plt.plot( df[colnames[0]].values
63 , df[colname].values / df[colnames[1]].values
67plt.xticks(fontsize = fontsize)
68plt.yticks(fontsize = fontsize)
69ax.set_xlabel(colnames[0], fontsize = fontsize)
70ax.set_ylabel(
"Runtime compared to {}".format(colnames[1]), fontsize = fontsize)
71ax.set_title(
"Runtime Ratio Comparison. Lower means faster.\nLower than 1 means faster than {}().".format(colnames[1]), fontsize = fontsize)
75plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
76ax.tick_params(axis =
"y", which =
"minor")
77ax.tick_params(axis =
"x", which =
"minor")
78ax.legend ( colnames[1:]
85plt.savefig(
"benchmark." + dirname +
".runtime.ratio.png")
Visualization of the benchmark output ⛓
- Todo:
- High Priority: A benchmark comparing the performance of the two computational algorithms above should be implemented and gauge the impact of row vs. column major access pattern.
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, Friday 1:54 AM, April 21, 2017, Institute for Computational Engineering and Sciences (ICES), The University of Texas, Austin, TX