7 real(RKG),
parameter :: EPS
= sqrt(
epsilon(
0._RKG))
8 real(RKG),
parameter :: LARGE
= sqrt(
huge(
0._RKG))
16 use auxil_mod,
only: RKG
21 public :: obs_type, posuniv_type, stat_type, data_type
23 type,
abstract :: obs_type
26 type,
extends(obs_type) :: univ_type
30 type,
extends(univ_type) :: posuniv_type
34 interface posuniv_type
35 module procedure :: posuniv_typer
44 type(stat_type) :: stat
45 class(obs_type),
allocatable :: obs(:)
49 module procedure :: data_typer
54 function posuniv_typer(val)
result(self)
55 real(RKG),
intent(in),
contiguous :: val(:)
56 type(posuniv_type),
allocatable :: self(:)
58 allocate(self(
size(val)))
59 do concurrent(iobs
= 1 :
size(val))
60 self(iobs)
%log
= log(val(iobs))
61 self(iobs)
%val
= val(iobs)
65 function data_typer(obs, stat)
result(self)
66 class(obs_type),
intent(in),
contiguous :: obs(:)
67 type(stat_type),
intent(in) :: stat
68 type(data_type) :: self
70 self
%nobs
= size(obs,
1, IK)
71 allocate(self
%obs,
source = obs)
81 use auxil_mod,
only: RKG
86 real(RKG),
allocatable :: lower(:)
87 real(RKG),
allocatable :: upper(:)
89 character(
63, SK),
allocatable :: names(:)
99 use data_mod,
only: obs_type
100 use domain_mod,
only: domain_type
101 use auxil_mod,
only: RKG
105 type(domain_type) :: domain
109 procedure(getParam_proc),
deferred :: getParam
110 procedure(setParam_proc),
deferred :: setParam
111 procedure(getLogPDF_proc),
deferred :: getLogPDF
115 class(model_type),
allocatable :: model
119 module procedure :: con_typer
123 function getParam_proc(self)
result(param)
125 class(model_type),
intent(in) :: self
126 real(RKG) :: param(self%npar)
128 subroutine setParam_proc(self, param)
130 class(model_type),
intent(inout) :: self
131 real(RKG),
intent(in),
contiguous :: param(:)
133 impure elemental function getLogPDF_proc(self, obs)
result(logPDF)
135 class(model_type),
intent(in) :: self
136 class(obs_type),
intent(in) :: obs
143 pure elemental function con_typer(model)
result(self)
144 class(model_type),
intent(in) :: model
145 type(con_type) :: self
155 use model_mod,
only: domain_type, model_type, IK
156 use auxil_mod,
only: RKG, LARGE
159 type,
extends(model_type) :: lognorm_type
160 real(RKG),
private :: avg, invstd, loginvstd
162 procedure :: getParam, setParam, getLogPDF
165 interface lognorm_type
166 module procedure :: lognorm_typer
171 function lognorm_typer(param, domain)
result(self)
172 real(RKG),
intent(in),
contiguous,
optional :: param(:)
173 type(domain_type),
intent(in),
optional :: domain
174 type(lognorm_type) :: self
176 if (
present(domain))
then
179 self
%domain
= domain_type([
-LARGE,
-LARGE], [
+LARGE,
+LARGE], [
character(
63) ::
"lognorm_avg",
"lognorm_logstd"])
181 if (
present(param))
call self
%setParam(param)
184 subroutine setParam(self, param)
186 class(lognorm_type),
intent(inout) :: self
187 real(RKG),
intent(in),
contiguous :: param(:)
188 if (
size(param)
/= self
%npar)
error stop "`lognorm_type%setParam()` takes a vector of two parameters representing `avg` and `logstd`."
189 self
%invstd
= exp(
-param(
2))
190 self
%loginvstd
= -param(
2)
194 function getParam(self)
result(param)
195 class(lognorm_type),
intent(in) :: self
196 real(RKG) :: param(self%npar)
197 param
= [self
%avg,
-self
%loginvstd]
200 impure elemental function getLogPDF(self, obs)
result(logPDF)
202 use data_mod,
only: obs_type, posuniv_type
203 class(lognorm_type),
intent(in) :: self
204 class(obs_type), intent(in):: obs
207 type is (posuniv_type)
208 call setNormLogPDF(logPDF, obs
%log, mu
= self
%avg, invSigma
= self
%invstd, logInvSigma
= self
%loginvstd)
210 error stop "Unrecognized data."
214end module lognorm_mod
220 use auxil_mod,
only: RKG, LARGE
221 use model_mod,
only: domain_type, model_type
224 type,
extends(model_type) :: flatPoweto_type
225 real(RKG),
private :: logbreak, alpha, break, alphap1, logPDFNF, limx(
2), loglimx(
2)
227 procedure,
pass :: getParam, setParam, getLogPDF
230 interface flatPoweto_type
231 module procedure :: flatPoweto_typer
236 function flatPoweto_typer(limx, param, domain)
result(self)
237 real(RKG),
intent(in),
contiguous,
optional :: param(:)
238 type(domain_type),
intent(in),
optional :: domain
239 real(RKG),
intent(in),
contiguous :: limx(:)
240 character(:),
allocatable :: msg
241 type(flatPoweto_type) :: self
244 self
%loglimx
= log(limx)
245 if (
present(domain))
then
248 self
%domain
= domain_type([
-LARGE,
-LARGE], [
+LARGE,
+LARGE], [
character(
63) ::
"flatPoweto_logbreak",
"flatPoweto_alpha"])
250 if (
present(param))
call self
%setParam(param)
253 function getParam(self)
result(param)
254 class(flatPoweto_type),
intent(in) :: self
255 real(RKG) :: param(self%npar)
256 param
= [self
%logbreak, self
%alpha]
259 subroutine setParam(self, param)
265 real(RKG),
intent(in),
contiguous :: param(:)
266 class(flatPoweto_type),
intent(inout) :: self
267 real(RKG) :: logModelInt1, logModelInt2, small, large
268 if (
size(param)
/= self
%npar)
error stop "`lognorm_type%setParam()` takes a vector of two parameters representing `avg` and `logstd`."
269 self
%alpha
= param(
2)
270 self
%logbreak
= param(
1)
271 self
%alphap1
= self
%alpha
+ 1
272 self
%break
= exp(self
%logbreak)
273 logModelInt1
= log(self
%break
- self
%limx(
1))
274 if (self
%alphap1
< 0._RKG)
then
275 logModelInt2
= -log(
-self
%alphap1)
276 large
= self
%alphap1
* self
%logbreak
277 small
= self
%alphap1
* self
%loglimx(
2)
278 elseif (
0._RKG < self
%alphap1)
then
279 logModelInt2
= -log(self
%alphap1)
280 small
= self
%alphap1
* self
%logbreak
281 large
= self
%alphap1
* self
%loglimx(
2)
283 logModelInt2
= 0._RKG
284 small
= self
%logbreak
285 large
= self
%loglimx(
2)
287 logModelInt2
= logModelInt2
- self
%alpha
* self
%logbreak
+ getLogSubExp(smaller
= small, larger
= large)
291 pure elemental function getLogPDF(self, obs)
result(logPDF)
292 use data_mod,
only: obs_type, posuniv_type
293 class(flatPoweto_type),
intent(in) :: self
294 class(obs_type),
intent(in) :: obs
297 type is (posuniv_type)
298 if (obs
%log
< self
%logbreak)
then
301 logPDF
= self
%alphap1
* obs
%log
- self
%alpha
* self
%logbreak
303 logPDF
= logPDF
+ self
%logPDFNF
305 error stop "Unrecognized data."
309end module flatPoweto_mod
313module flatPowetoTapered_mod
315 use auxil_mod,
only: RKG, LARGE
316 use model_mod,
only: domain_type, model_type
319 type,
extends(model_type) :: flatPowetoTapered_type
320 real(RKG),
private :: logbreak, alpha, beta, break, alphap1, logPDFNF, limx(
2), loglimx(
2)
322 procedure,
private :: getLogUDF
323 procedure :: getParam, setParam, getLogPDF
326 interface flatPowetoTapered_type
327 module procedure :: flatPowetoTapered_typer
332 function flatPowetoTapered_typer(limx, param, domain)
result(self)
333 real(RKG),
intent(in),
contiguous,
optional :: param(:)
334 type(domain_type),
intent(in),
optional :: domain
335 real(RKG),
intent(in),
contiguous :: limx(:)
336 type(flatPowetoTapered_type) :: self
339 self
%loglimx
= log(limx)
340 if (
present(domain))
then
343 self
%domain
= domain_type([
-LARGE,
-LARGE,
-LARGE], [
+LARGE,
+LARGE,
+LARGE], [
character(
63) ::
"flatPowetoTapered_logbreak",
"flatPowetoTapered_alpha",
"flatPowetoTapered_beta"])
345 if (
present(param))
call self
%setParam(param)
348 function getParam(self)
result(param)
349 class(flatPowetoTapered_type),
intent(in) :: self
350 real(RKG) :: param(self%npar)
351 param
= [self
%logbreak, self
%alpha, self
%beta]
354 subroutine setParam(self, param)
360 class(flatPowetoTapered_type),
intent(inout) :: self
361 real(RKG),
intent(in),
contiguous :: param(:)
362 real(RKG) :: logModelInt1, logModelInt2, logNF
363 character(
255, SK) :: msg
364 logical(LK) :: failed
365 if (
size(param)
/= self
%npar)
error stop "`flatPowetoFlatPowetoTapered_type` takes a vector of three parameters"&
366 //"representing `logbreak`, `alpha`, `beta`. size(param), npar = "//getStr([
size(param,
1,
IK), self
%npar])
367 self
%logbreak
= param(
1)
368 self
%alpha
= param(
2)
370 self
%alphap1
= self
%alpha
+ 1
371 self
%break
= exp(self
%logbreak)
372 logModelInt1
= log(self
%break
- self
%limx(
1))
373 logNF
= max(self
%logbreak, self
%alphap1
* self
%loglimx(
2)
- self
%alpha
* self
%logbreak
- self
%beta
* (self
%limx(
2)
- self
%break))
374 failed
= isFailedQuad(getDensity, lb
= self
%logbreak, ub
= self
%loglimx(
2), help
= weps, integral
= logModelInt2, msg
= msg)
377 self
%logPDFNF
= sqrt(
huge(self
%logPDFNF))
379 logModelInt2
= log(logModelInt2)
+ logNF
383 function getDensity(logx)
result(density)
384 real(RKG),
intent(in) :: logx
386 density
= exp(self
%getLogUDF(logx,
exp(logx))
- logNF)
390 pure elemental function getLogPDF(self, obs)
result(logPDF)
391 use data_mod,
only: obs_type, posuniv_type
392 class(flatPowetoTapered_type),
intent(in) :: self
393 class(obs_type), intent(in):: obs
396 type is (posuniv_type)
397 if (obs
%log
< self
%logbreak)
then
400 logPDF
= self
%getLogUDF(obs
%log, obs
%val)
402 logPDF
= logPDF
+ self
%logPDFNF
404 error stop "Unrecognized data."
408 pure elemental function getLogUDF(self, logx, x)
result(logUDF)
409 class(flatPowetoTapered_type),
intent(in) :: self
410 real(RKG),
intent(in) :: logx, x
412 logUDF
= self
%alphap1
* logx
- self
%alpha
* self
%logbreak
- self
%beta
* (x
- self
%break)
415end module flatPowetoTapered_mod
425 use model_mod,
only: domain_type, model_type, con_type
426 use auxil_mod,
only: RKG, EPS, LARGE
427 use data_mod,
only: obs_type
432 type(con_type),
allocatable :: comp(:)
433 real(RKG),
allocatable :: logfrac(:)
434 integer(IK),
private :: ncomp
436 procedure :: getParam, setParam, getLogPDF
439 interface mixture_type
440 module procedure :: mixture_typer
445 function mixture_typer(comp, param)
result(self)
446 type(con_type),
intent(in),
contiguous :: comp(:)
447 real(RKG),
intent(in),
contiguous,
optional :: param(:)
448 character(:, SK),
allocatable :: prefix
449 integer(IK) :: icomp, lpar, upar, ipar
450 type(mixture_type) :: self
451 self
%ncomp
= size(comp,
1, IK)
452 allocate(self
%comp,
source = comp)
454 self
%npar
= sum([(self
%comp(icomp)
%model
%npar, icomp
= 1, self
%ncomp)])
+ self
%ncomp
- 1
459 do icomp
= 1, self
%ncomp
- 1
460 upar
= lpar
+ self
%comp(icomp)
%model
%npar
461 prefix
= SK_
"comp"//getStr(icomp)
//SK_
"_"
462 self
%domain
%lower(lpar
+ 1 : upar
+ 1)
= [self
%comp(icomp)
%model
%domain
%lower,
-LARGE]
463 self
%domain
%upper(lpar
+ 1 : upar
+ 1)
= [self
%comp(icomp)
%model
%domain
%upper,
+LARGE]
464 self
%domain
%names(lpar
+ 1 : upar
+ 1)
= [
character(
63, SK) :: prefix
//self%comp(icomp)%model%domain%names, prefix
//SK_
"fisherz(frac)"]
467 prefix
= SK_
"comp"//getStr(icomp)
//SK_
"_"
468 self
%domain
%lower(lpar
+ 1 :)
= self
%comp(icomp)
%model
%domain
%lower
469 self
%domain
%upper(lpar
+ 1 :)
= self
%comp(icomp)
%model
%domain
%upper
470 self
%domain
%names(lpar
+ 1 :)
= prefix
//self
%comp(icomp)
%model
%domain
%names
471 if (
present(param))
call self
%setParam(param)
474 subroutine setParam(self, param)
476 class(mixture_type),
intent(inout) :: self
477 real(RKG),
intent(in),
contiguous :: param(:)
478 integer(IK) :: icomp, lpar, upar
479 real(RKG) :: sumfrac, mixfrac
480 if (
size(param)
/= self
%npar)
error stop "@mixture_type%setParam(): The condition `size(param) == self%npar` must hold. size(param), self%npar = "//getStr([
size(param), self
%npar])
483 do icomp
= 1, self
%ncomp
- 1
484 upar
= lpar
+ self
%comp(icomp)
%model
%npar
485 call self
%comp(icomp)
%model
%setParam(param(lpar
+ 1 : upar))
487 self
%logfrac(icomp)
= log(mixfrac)
488 sumfrac
= sumfrac
+ mixfrac
491 self
%logfrac(icomp)
= log(
1 - sumfrac)
492 call self
%comp(icomp)
%model
%setParam(param(lpar
+ 1 :))
493 if (
.not. abs(
1 - sum(
exp(self
%logfrac)))
< EPS)
error stop "The condition `sum(exp(self%logfrac)) == 1` must hold. self%logfrac = "//getStr(self
%logfrac)
496 function getParam(self)
result(param)
498 class(mixture_type),
intent(in) :: self
499 integer(IK) :: icomp, lpar, upar
500 real(RKG) :: param(self%npar)
501 param
= [(self
%comp(icomp)
%model
%getParam(),
getFisher(
exp(self
%logfrac(icomp)),
0._RKG,
1._RKG), icomp
= 1, self
%ncomp
- 1), self
%comp(self
%ncomp)
%model
%getParam()]
504 impure elemental function getLogPDF(self, obs)
result(logPDF)
505 class(mixture_type),
intent(in) :: self
506 real(RKG) :: logPDFS(self%ncomp), maxLogPDF
507 class(obs_type),
intent(in) :: obs
510 maxLogPDF
= -huge(
0._RKG)
511 do icomp
= 1, self
%ncomp
512 logPDFS(icomp)
= self
%logfrac(icomp)
+ self
%comp(icomp)
%model
%getLogPDF(obs)
513 maxLogPDF
= max(maxLogPDF, logPDFS(icomp))
518end module mixture_mod
524 use auxil_mod,
only: RKG
527 use model_mod,
only: model_type
528 use data_mod,
only: data_type
536 real(RKG),
allocatable :: param(:)
541 type(best_type) :: best
542 type(data_type) :: data
543 class(model_type),
allocatable :: model
544 class(sampler_type),
allocatable :: sampler
546 procedure,
pass :: run
551 module procedure :: fit_typer
556 function fit_typer(data, model, sampler)
result(self)
557 class(sampler_type),
intent(in),
optional :: sampler
558 class(model_type),
intent(in) :: model
559 type(data_type),
intent(in) :: data
560 type(fit_type) :: self
561 if (
present(sampler))
then
562 self
%sampler
= sampler
575 class(fit_type),
intent(inout) :: self
576 type(err_type) :: err
578 if (
.not. allocated(self
%sampler
%outputStatus)) self
%sampler
%outputStatus
= "retry"
579 if (
.not. allocated(self
%sampler
%domainAxisName)) self
%sampler
%domainAxisName
= self
%model
%domain
%names
580 if (
.not. allocated(self
%sampler
%domainCubeLimitLower)) self
%sampler
%domainCubeLimitLower
= self
%model
%domain
%lower
581 if (
.not. allocated(self
%sampler
%domainCubeLimitUpper)) self
%sampler
%domainCubeLimitUpper
= self
%model
%domain
%upper
583 select type (sampler
=> self
%sampler)
585 if (
.not. allocated(sampler
%outputChainSize)) sampler
%outputChainSize
= 30000
586 if (
.not. allocated(sampler
%proposalScale)) sampler
%proposalScale
= "gelman"
587 if (
.not. allocated(sampler
%proposalStart)) sampler
%proposalStart
= self
%model
%getParam()
590 if (err
%occurred)
error stop "sampler failed: "//err
%msg
600 integer(IK) :: stat, ibest
601 type(css_type),
allocatable :: path(:)
602 real(RKG),
allocatable :: logx(:), logPDF(:)
603 real(RKG),
allocatable :: table(:,:), state(:)
605 if (
getImageID()
== 1)
write(
*,
"(A)")
"Searching for files: "//self
%sampler
%outputFileName
//SK_
"*"
606 path
= glob(self
%sampler
%outputFileName
//SK_
"*")
607 if (
size(path)
== 0)
error stop "There is no sample file in the output folder."
610 if (stat
/= 0)
error stop "Failed to read output sample."
611 ibest
= maxloc(table(:,
1),
dim = 1_IK)
612 self
%best
%loglike
= table(ibest,
1)
613 self
%best
%param
= table(ibest,
2:)
614 self
%bic
= self
%model
%npar
* log(
real(self
%data
%nobs, RKG))
- 2 * self
%best
%loglike
620 function getLogLike(param)
result(logLike)
621 real(RKG),
intent(in),
contiguous :: param(:)
623 call self
%model
%setParam(param)
624 loglike
= sum(self
%model
%getLogPDF(self
%data
%obs))
636 use fit_mod,
only: fit_type
637 use model_mod,
only: con_type
638 use auxil_mod,
only: RKG, LARGE
639 use lognorm_mod,
only: lognorm_type
640 use mixture_mod,
only: mixture_type
643 use flatPoweto_mod,
only: flatPoweto_type
644 use data_mod,
only: posuniv_type, data_type, stat_type
645 use flatPowetoTapered_mod,
only: flatPowetoTapered_type
648 integer(IK) :: imodel, stat
649 type(paradram_type) :: sampler
650 type(data_type) :: data, pred
651 type(display_type) :: disp
652 type(fit_type) :: fit
660 real(RKG),
allocatable :: table(:,:)
661 character(
255, SK) :: iomsg
663 if (stat
/= 0)
error stop "Failed to read data: "//trim(iomsg)
664 associate(val
=> table(:,
4))
665 data = data_type(obs
= posuniv_type(val),
stat = stat_type([
minval(val),
maxval(val)]))
674 sampler
%parallelismMpiFinalizeEnabled
= .false.
676 if (imodel
== 1)
then
677 sampler
%outputFileName
= "./mixLogNormLogNorm"
678 sampler
%proposalStart
= [
-.
2,
0.4, .
4,
3.6,
-0.2]
679 sampler
%domainCubeLimitLower
= [
real(RKG) ::
-LARGE,
-LARGE,
-LARGE,
log(
3.),
-LARGE]
680 sampler
%domainCubeLimitUpper
= [
real(RKG) ::
log(
3.),
+LARGE,
+LARGE,
+LARGE,
+LARGE]
682 sampler
%proposalCov
= reshape (
&
683 [
1.6E-2,
5.5E-3,
4.1E-3,
3.9E-3,
-2.5E-3 &
684 ,
5.5E-3,
3.1E-3,
1.7E-3,
1.4E-3,
-9.5E-4 &
685 ,
4.1E-3,
1.7E-3,
1.9E-3,
1.2E-3,
-8.8E-4 &
686 ,
3.9E-3,
1.4E-3,
1.2E-3,
2.0E-3,
-8.9E-4 &
687 ,
-2.5E-3,
-9.5E-4,
-8.8E-4,
-8.9E-4,
1.0E-3 &
688 ], shape
= [
size(sampler
%proposalStart),
size(sampler
%proposalStart)])
689 fit
= fit_type(data, mixture_type([con_type(lognorm_type()), con_type(lognorm_type())]), sampler)
690 elseif (imodel
== 2)
then
691 sampler
%proposalScale
= "0.1 * gelman"
692 sampler
%outputFileName
= "./mixFlatPowetoFlatPowetoTapered"
693 sampler
%proposalStart
= [
log(.
37),
-1.4, .
3,
log(
21.3),
-.
5,
0.0156]
694 sampler
%proposalCov
= reshape (
&
695 [
1.E-2,
-3.E-3,
1.E-2,
-7.E-3,
-1.E-4,
-6.E-4 &
696 ,
-3.E-3,
3.E-3,
-4.E-3,
6.E-3,
1.E-4,
1.E-3 &
697 ,
1.E-2,
-4.E-3,
1.E-1,
-7.E-2,
-8.E-4,
3.E-4 &
698 ,
-7.E-3,
6.E-3,
-7.E-2,
7.E-2,
8.E-4,
3.E-3 &
699 ,
-1.E-4,
1.E-4,
-8.E-4,
8.E-4,
1.E-5,
6.E-5 &
700 ,
-6.E-4,
1.E-3,
3.E-4,
3.E-3,
6.E-5,
2.E-3 &
701 ], shape
= [
size(sampler
%proposalStart),
size(sampler
%proposalStart)])
702 fit
= fit_type(data, mixture_type([con_type(flatPoweto_type(data
%stat
%lim)), con_type(flatPowetoTapered_type(data
%stat
%lim))]), sampler)
703 elseif (imodel
== 3)
then
704 sampler
%proposalScale
= "0.1 * gelman"
705 sampler
%outputFileName
= "./mixLogNormFlatPowetoTapered"
706 sampler
%proposalStart
= [
-.
2,
0.4, .
3,
log(
21.3),
-.
5,
0.0156]
707 fit
= fit_type(data, mixture_type([con_type(lognorm_type()), con_type(flatPowetoTapered_type(data
%stat
%lim))]), sampler)
708 elseif (imodel
== 4)
then
709 sampler
%proposalScale
= "0.1 * gelman"
710 sampler
%outputFileName
= "./mixFlatPowetoLogNorm"
711 sampler
%proposalStart
= [
log(.
37),
-1.4, .
3,
3.6,
-0.2]
712 fit
= fit_type(data, mixture_type([con_type(flatPoweto_type(data
%stat
%lim)), con_type(lognorm_type())]), sampler)
714 sampler
%outputFileName
= "./logNorm"
715 fit
= fit_type(data, lognorm_type([
real(RKG) ::
1,
1]), sampler)
720 call disp%show(css_type(fit
%sampler
%domainAxisName))
723 call disp%show(
"[fit%best%loglike, fit%bic]")
724 call disp%show( [fit
%best
%loglike, fit
%bic] )
729 associate(val
=> getLogSpace(logx1
= log(data
%stat
%lim(
1)), logx2
= log(data
%stat
%lim(
2)), count
= 1000_IK))
730 pred
= data_type(obs
= posuniv_type(val),
stat = stat_type([
minval(val),
maxval(val)]))
731 call fit
%model
%setParam(fit
%best
%param)
732 stat
= getErrTableWrite(fit
%sampler
%outputFileName
//SK_
".hist",
reshape([
log(val), fit
%model
%getLogPDF(pred
%obs)], [
size(val),
2]), header
= SK_
"logx,logPDF")
733 if (stat
/= 0)
error stop "Failed to write the histogram visualization data."
Allocate or resize (shrink or expand) an input allocatable scalar string or array of rank 1....
Generate count evenly-logarithmically-spaced points over the interval [base**logx1,...
Generate the natural logarithm of probability density function (PDF) of the univariate Normal distrib...
Generate and return the iostat code resulting from reading the contents of the specified file or unit...
Generate and return the iostat code resulting from writing the input table of rank 1 or 2 to the spec...
This is a generic method of the derived type display_type with pass attribute.
Generate and return the inverse Fisher transformation of the input Fisher z value.
Generate and return the Fisher transformation of the input Fisher z value.
Generate and return the logarithm of the sum of the exponential of two input logarithmic values robus...
Generate and return the natural logarithm of the subtraction of the exponential of the smaller input ...
Generate and return the natural logarithm of the sum of the exponential of the input array robustly (...
Generate and return an array of size two containing the minimum and maximum of the two input values i...
Compute the 1D integral of the input scalar (potentially singular) integrand getFunc on a finite or s...
Compute the 1D integral of the input scalar (potentially singular) integrand getFunc on a finite or s...
Generate and return the conversion of the input value to an output Fortran string,...
This module contains procedures and generic interfaces for resizing allocatable arrays of various typ...
This module contains procedures and generic interfaces for generating arrays with linear or logarithm...
This module contains classes and procedures for computing various statistical quantities related to t...
This module contains procedures and generic interfaces for evaluating the Fisher transformation and i...
This module contains procedures and generic interfaces for adding two real or complex values without ...
This module contains procedures and generic interfaces for subtracting two real or complex values wit...
This module contains the procedures and interfaces for computing the natural logarithm of the sum of ...
This module contains procedures and generic interfaces for finding the minimum and maximum of two inp...
This module contains procedures and generic interfaces for facilitating parallel computations or comp...
integer(IK) function getImageID()
Generate and return the ID of the current Coarray image / MPI process / OpenMP thread,...
This module contains classes and procedures for non-adaptive and adaptive global numerical quadrature...
type(weps_type), parameter weps
The scalar constant object of type weps_type that indicates the use of Epsilon extrapolation method o...
This module contains the generic procedures for converting values of different types and kinds to For...
This is the abstract derived type for creating objects of class model_type that contain the character...
This is a derived type for constructing objects containing the optional simulation properties of the ...
3import matplotlib.pyplot
as plt
11plt.rcParams[
'text.usetex'] =
True
14examname = os.path.basename(os.getcwd())
20files = glob.glob(
"./*_chain.txt")
24 basename = file.split(
"_")[0]
25 df = pd.read_csv(file, delimiter =
",")
26 if "_chain.txt" in file:
28 elif "_sample.txt" in file:
31 sys.exit(
"Unrecognized simulation output file: " + file)
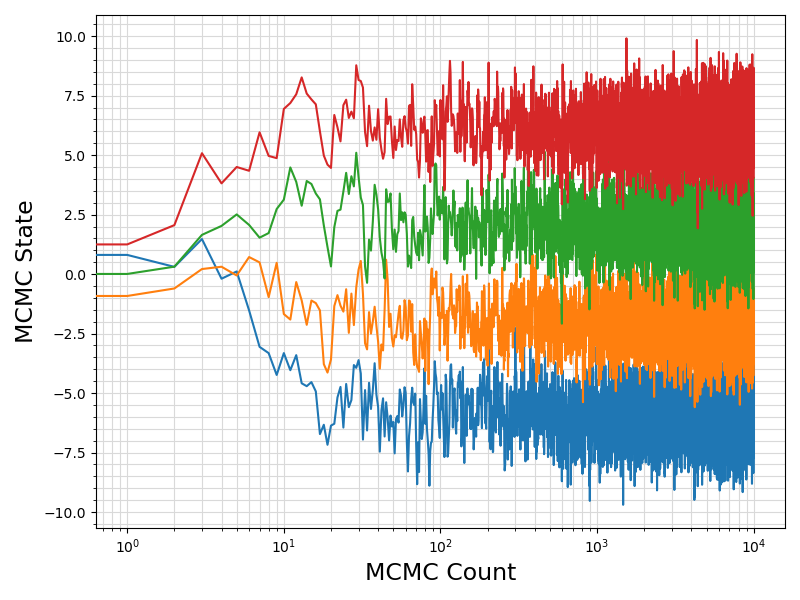
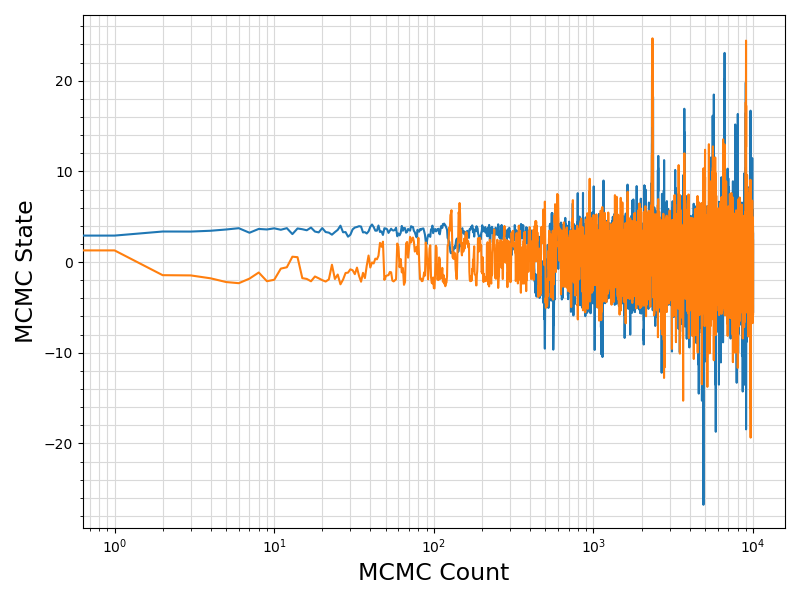
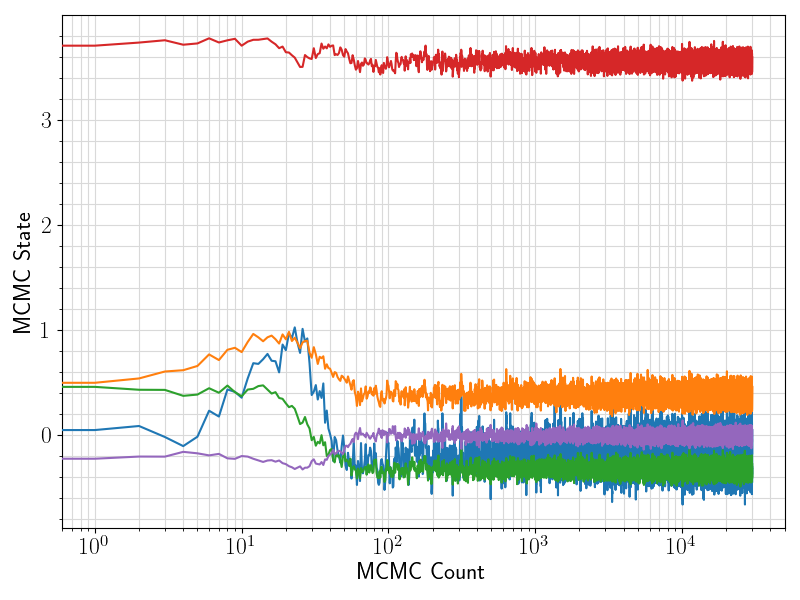
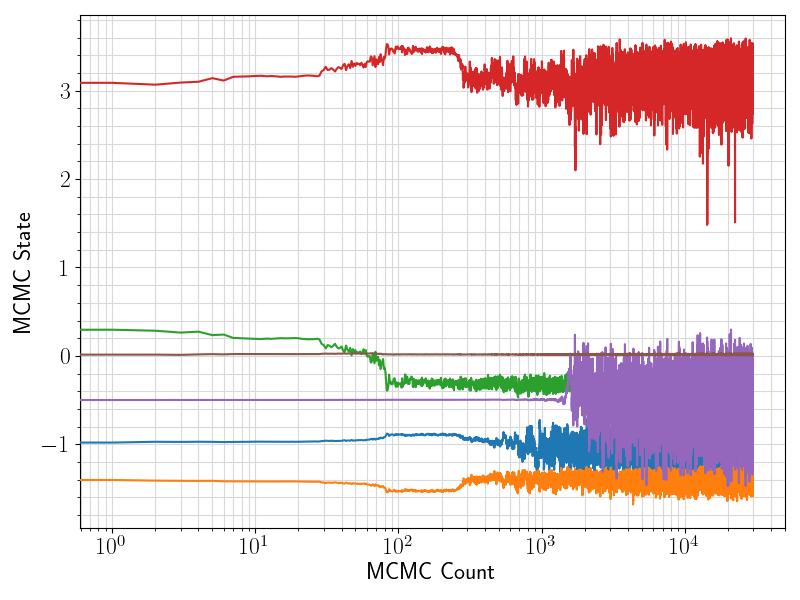
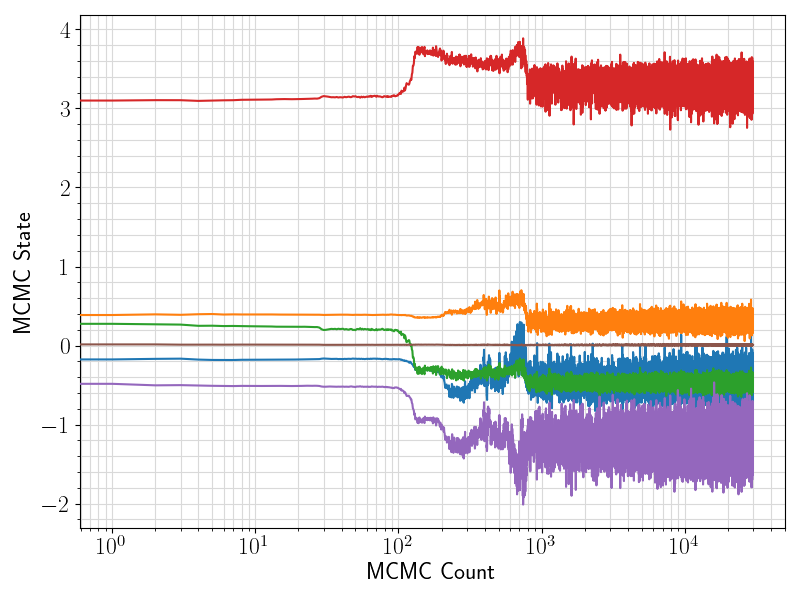
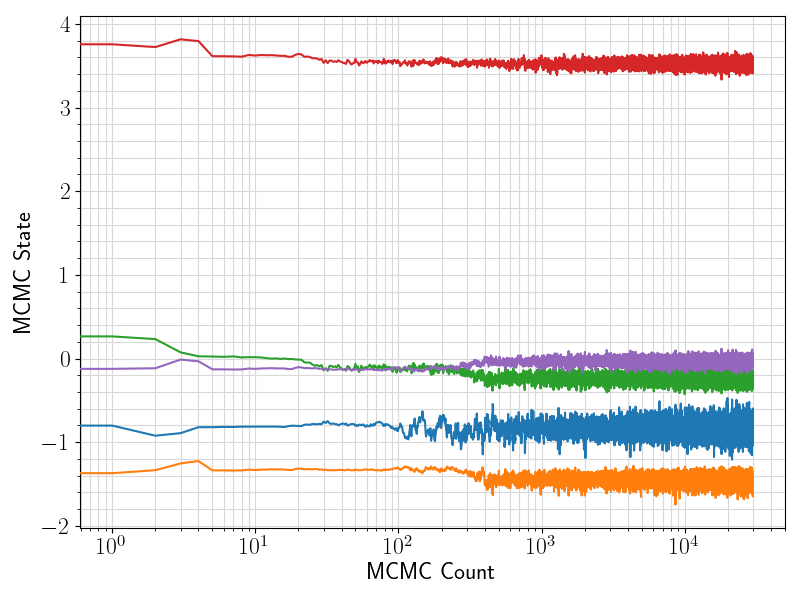
36 fig = plt.figure(figsize = (8, 6))
37 ax = plt.subplot(1,1,1)
38 ax.plot ( range(len(df.values[:, 0]))
39 , df.values[:, sindex:]
44 ax.set_xlabel(
"MCMC Count", fontsize = 17)
45 ax.set_ylabel(
"MCMC State", fontsize = 17)
46 ax.tick_params(axis =
"x", which =
"major", labelsize = fontsize)
47 ax.tick_params(axis =
"y", which =
"major", labelsize = fontsize)
48 plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
52 plt.savefig(basename +
".traceplot.png")
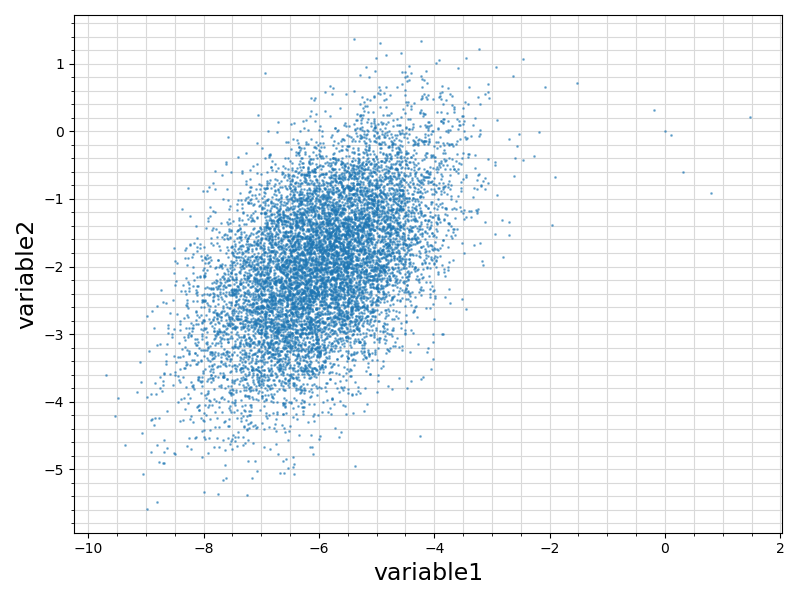
56 if len(df.values[1, sindex:]) == 2:
58 fig = plt.figure(figsize = (8, 6))
59 ax = plt.subplot(1,1,1)
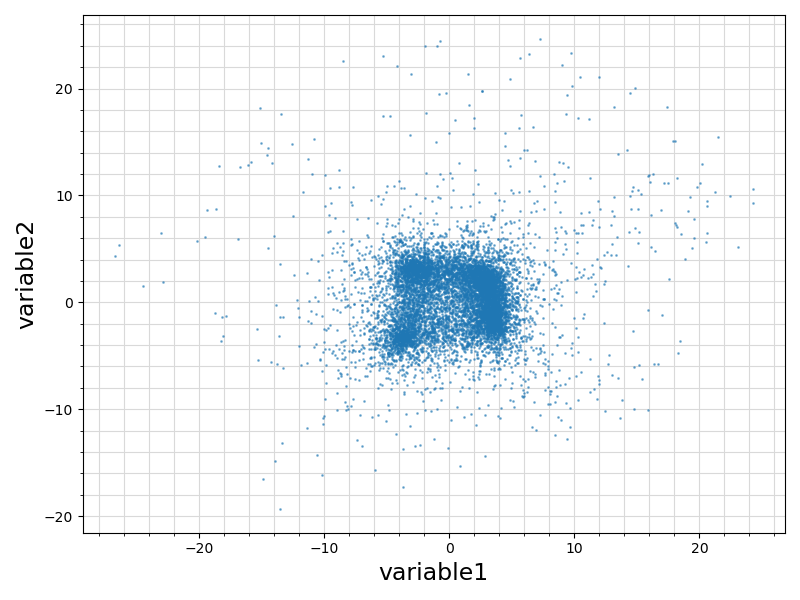
60 ax.scatter ( df.values[:, sindex]
61 , df.values[:, sindex + 1]
67 ax.set_xlabel(df.columns.values[sindex], fontsize = 17)
68 ax.set_ylabel(df.columns.values[sindex + 1], fontsize = 17)
69 ax.tick_params(axis =
"x", which =
"major", labelsize = fontsize)
70 ax.tick_params(axis =
"y", which =
"major", labelsize = fontsize)
71 plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
75 plt.savefig(basename +
".scatterplot.png")
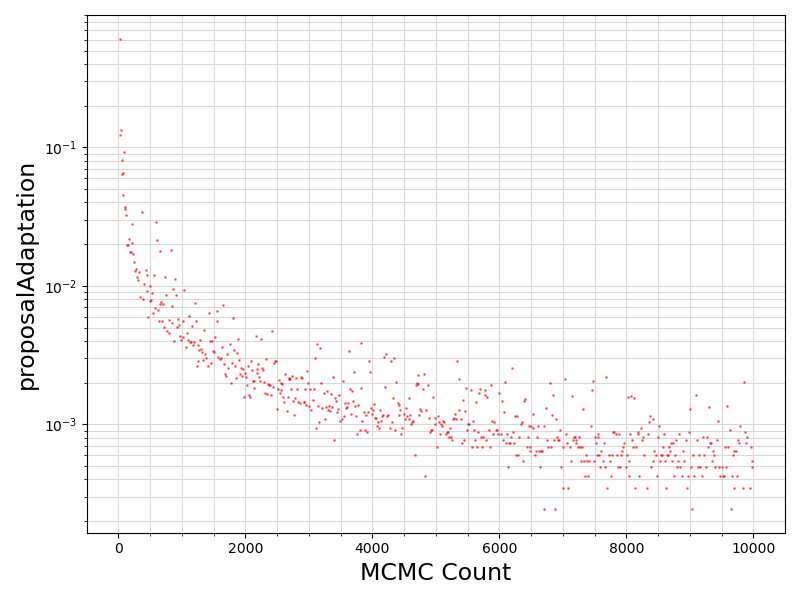
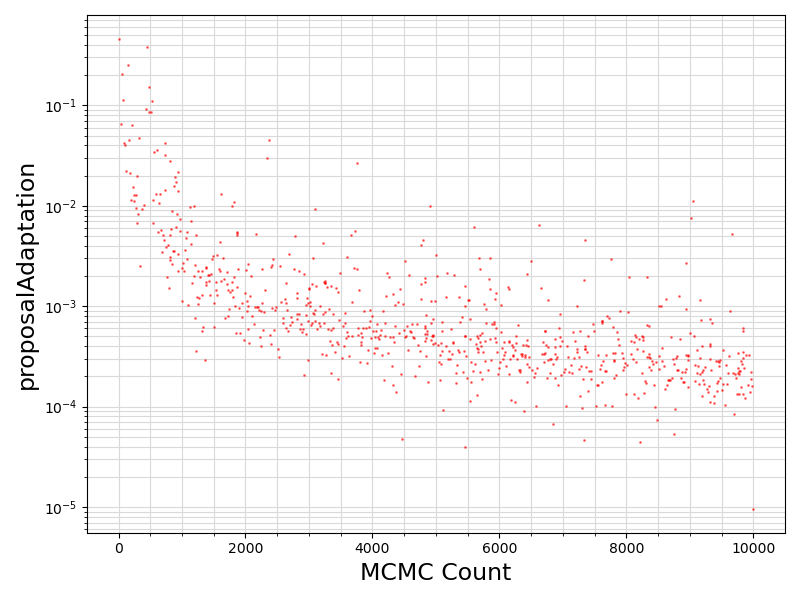
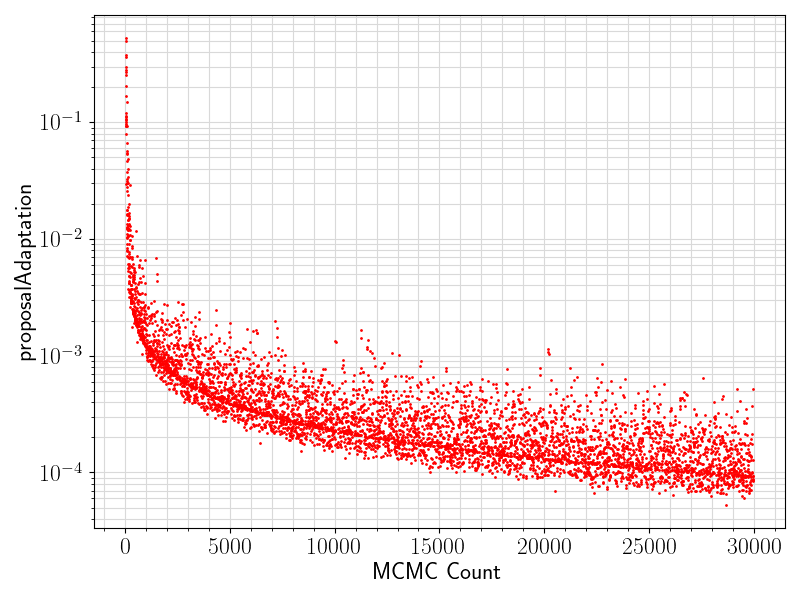
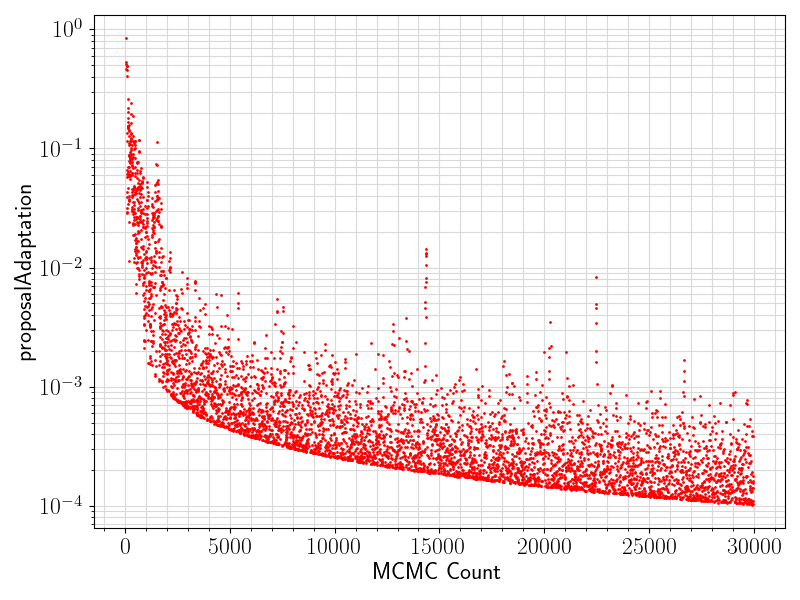
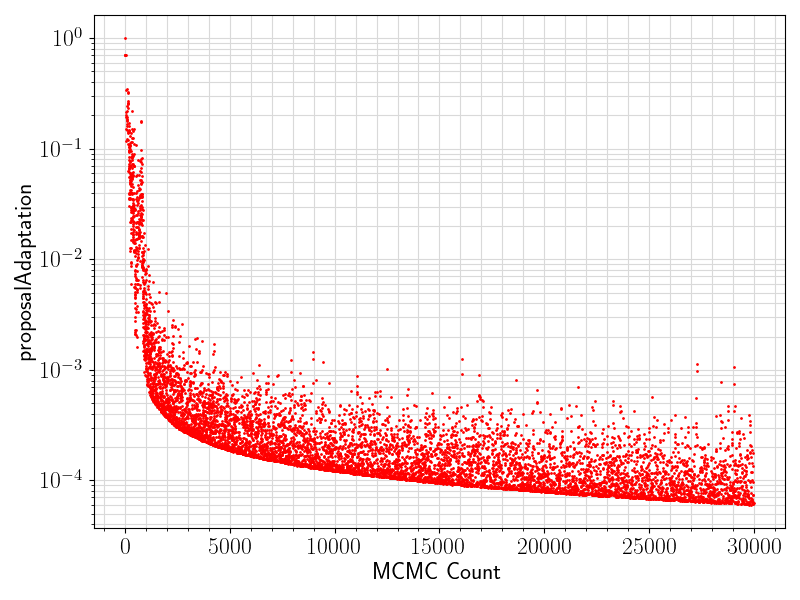
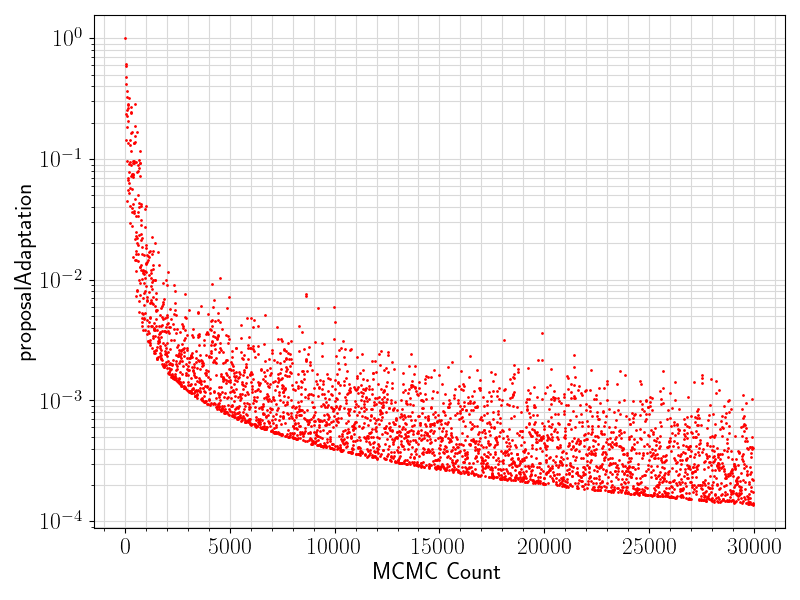
80 if "proposalAdaptation" in df.columns.values:
81 if any(df[
"proposalAdaptation"].values != 0):
82 fig = plt.figure(figsize = (8, 6))
83 ax = plt.subplot(1,1,1)
84 ax.scatter ( range(len(df.values[:, 0]))
85 , df[
"proposalAdaptation"].values
93 ax.set_xlabel(
"MCMC Count", fontsize = 17)
94 ax.tick_params(axis =
"x", which =
"major", labelsize = fontsize)
95 ax.tick_params(axis =
"y", which =
"major", labelsize = fontsize)
96 ax.set_ylabel(
"proposalAdaptation", fontsize = 17)
97 plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
101 plt.savefig(basename +
".proposalAdaptation.png")
113def mergeBins(counts, bins, threshold = 5):
116 halfway = int(np.ceil(len(bins)/2))
118 halfway = int(np.floor(len(bins)/2))
120 threshold = threshold
124 rightBins = [bins[-1]]
129 for i
in range(0 , halfway):
132 leftSumCounts += counts[i]
136 if leftSumCounts >= threshold:
138 leftCounts += [leftSumCounts]
140 leftBins += [bins[i+1]]
145 rightSumCounts += counts[-i-1]
146 if rightSumCounts >= threshold:
147 rightCounts += [rightSumCounts]
149 rightBins += [bins[-i-2]]
152 if len(rightCounts) != 0:
157 rightCounts += [counts[halfway]]
160 newCounts = leftCounts + rightCounts[-1::-1]
161 newBins = leftBins + rightBins[-1::-1]
163 origionaldNdT = np.array(counts) / np.array([bins[i + 1] - bins[i]
for i
in range(0, len(bins) - 1) ])
164 newdNdT = np.array(newCounts) / np.array([newBins[i + 1] - newBins[i]
for i
in range(0, len(newBins) - 1) ])
166 return np.array(newCounts), np.array(newBins), np.array(origionaldNdT), np.array(newdNdT)
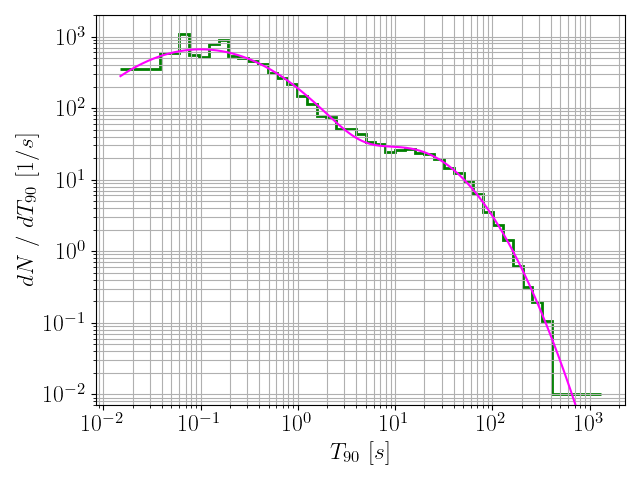
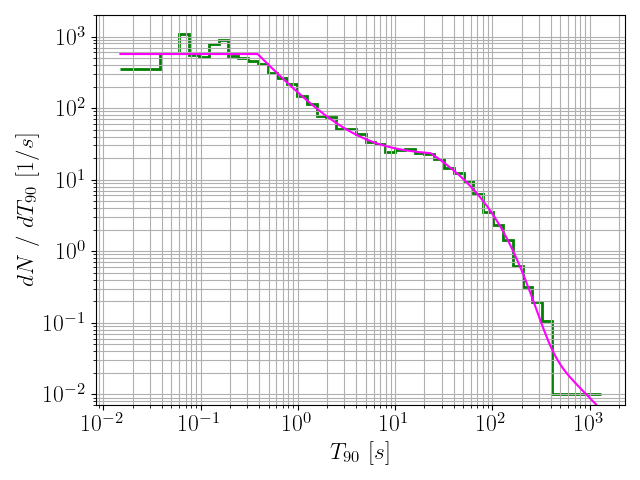
170data = pd.read_csv(
"data.csv", delimiter =
",")
171durs = data[
"T90"].values
174binning = 10**np.linspace(np.log10(min(durs)), np.log10(max(durs)), nbins)
175countsAndBins = np.histogram(durs, bins = binning)
176counts = countsAndBins[0]
177bins = countsAndBins[1]
179newCounts, newBins, origionaldNdT, dNdT = mergeBins(counts, binning)
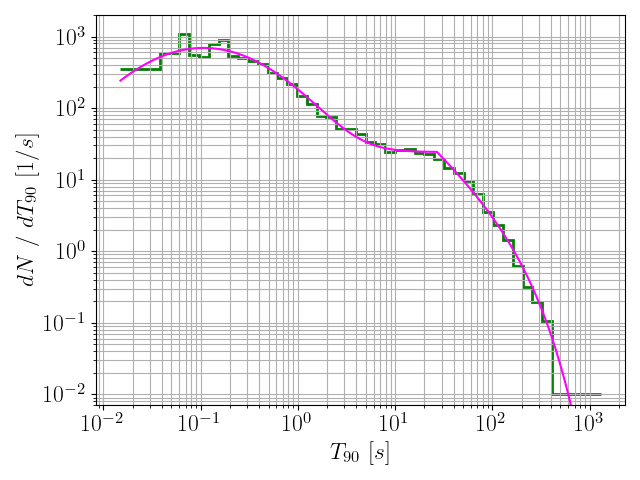
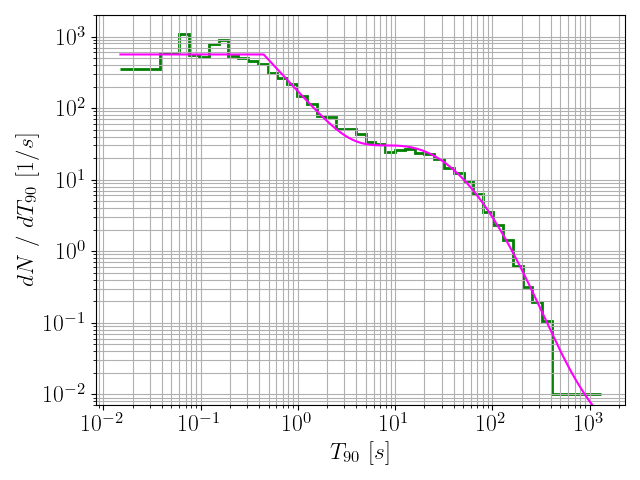
191histFiles = glob.glob(
"./*.hist")
193for histFile
in histFiles:
208 model = pd.read_csv(histFile, delimiter =
",")
209 x = np.exp(model[
"logx"].values)
210 y = len(durs) * np.exp(model[
"logPDF"].values) / x
211 plt.grid(which =
"both")
212 plt.plot(x, y, c =
"magenta", label = histFile)
213 plt.xlabel(
r"$T_{90} ~ [s]$", fontsize = fontsize)
214 plt.ylabel(
r"$dN ~ / ~ dT_{90} ~ [1 / s]$", fontsize = fontsize)
216 plt.xticks(size = fontsize)
217 plt.yticks(size = fontsize)
218 plt.ylim(7*10**-3, 2*10**3)
220 plt.savefig(histFile +
".png")