Compute and return an asymptotically optimal set of cluster centers for the input sample, cluster membership IDs, and sample distances-squared from their corresponding cluster centers.
More...
Compute and return an asymptotically optimal set of cluster centers for the input sample, cluster membership IDs, and sample distances-squared from their corresponding cluster centers.
In data mining, the k-means++ is an algorithm for choosing the initial values (or seeds) for the k-means clustering algorithm.
It was proposed in 2007 by David Arthur and Sergei Vassilvitskii, as an approximation algorithm for the NP-hard k-means problem.
It offers a way of avoiding the sometimes poor clustering found by the standard k-means algorithm.
Intuition
The intuition behind k-means++ is that spreading out the \(k\) initial cluster centers is a good thing:
The first cluster center is chosen uniformly at random from the data points that are being clustered, after which each subsequent cluster center is chosen from the remaining data points with probability proportional to its squared distance from the closest existing cluster center to the point.
Algorithm
The exact algorithm is as follows:
-
Choose one center uniformly at random among the data points.
-
For each data point \(x\) not chosen yet, compute \(D(x)\), the distance between \(x\) and the nearest center that has already been chosen.
-
Choose one new data point at random as a new center, using a weighted probability distribution where a point \(x\) is chosen with probability proportional to \(D(x)^2\).
-
Repeat Steps 2 and 3 until \(k\) centers have been chosen.
-
Now that the initial centers have been chosen, proceed using standard k-means clustering.
Performance improvements
The k-means++ seeding method yields considerable improvement in the final error of k-means algorithm.
Although the initial selection in the algorithm takes extra time, the k-means part itself converges very quickly after this seeding and thus the algorithm actually lowers the computation time.
Based on the original paper, the method yields typically 2-fold improvements in speed, and for certain datasets, close to 1000-fold improvements in error.
In these simulations the new method almost always performed at least as well as vanilla k-means in both speed and error.
- Note
- The metric used within this generic interface is the Euclidean distance.
- Parameters
-
| [in,out] | rng | : The input/output scalar that can be an object of,
-
type rngf_type, implying the use of intrinsic Fortran uniform RNG.
-
type xoshiro256ssw_type, implying the use of xoshiro256** uniform RNG.
|
| [out] | membership | : The output vector of shape (1:nsam) of type integer of default kind IK, containing the membership of each input sample in sample from its nearest cluster center, such that cluster(membership(i)) is the nearest cluster center to the ith sample sample(:, i) at a squared-distance of disq(i).
|
| [out] | disq | : The output vector of shape (1:nsam) of the same type and kind as the input argument sample, containing the Euclidean squared distance of each input sample in sample from its nearest cluster center.
|
| [in] | sample | : The input vector, or matrix of,
-
type
real of kind any supported by the processor (e.g., RK, RK32, RK64, or RK128),
containing the sample of nsam points in a ndim-dimensional space whose corresponding cluster centers must be computed.
-
If
sample is a vector of shape (1 : nsam) and center is a vector of shape (1 : ncls), then the input sample must be a collection of nsam points (in univariate space).
-
If
sample is a matrix of shape (1 : ndim, 1 : nsam) and center is a matrix of shape (1 : ndim, 1 : ncls), then the input sample must be a collection of nsam points (in ndim-dimensional space).
|
| [in] | ncls | : The input scalar of type integer of default kind IK, containing the number of the desired clusters to be identified in the sample.
(optional, default = ubound(center, 2). It must be present if and only if the output arguments center, size, and potential are all missing.) |
| [out] | center | : The output vector of shape (1:ncls) or matrix of shape (1 : ndim, 1 : ncls) of the same type and kind as the input argument sample, containing the set of ncls unique random cluster centers (centroids) selected from the input sample based on the computed memberships and minimum sample-cluster distances disq.
(optional. If missing, no cluster center information will be output.) |
| [out] | size | : The output vector of shape (1:ncls) type integer of default kind IK, containing the sizes (number of members) of the clusters with the corresponding centers output in the argument center.
(optional. If missing, no cluster size information will be output.) |
| [out] | potential | : The output vector of shape (1:ncls) of the same type and kind as the input argument sample, the ith element of which contains the sum of squared distances of all members of the ith cluster from the cluster center as output in the ith element of center.
(optional. If missing, no cluster potential information will be output.) |
Possible calling interfaces ⛓
call setKmeansPP(rng, membership(
1 : nsam) , disq(
1 : nsam), sample(
1 : ndim,
1 : nsam), ncls)
call setKmeansPP(rng, membership(
1 : nsam) , disq(
1 : nsam), sample(
1 : ndim,
1 : nsam), center(
1 : ndim,
1 : ncls),
size(
1 : ncls), potential(
1 : ncls))
Compute and return an asymptotically optimal set of cluster centers for the input sample,...
This module contains procedures and routines for the computing the Kmeans clustering of a given set o...
- Warning
- The condition
ubound(center, rank(center)) > 0 must hold for the corresponding input arguments.
The condition ubound(sample, rank(sample)) == ubound(disq, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) == ubound(size, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) == ubound(potential, 1) must hold for the corresponding input arguments.
The condition ubound(sample, rank(sample)) == ubound(membership, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) <= ubound(sample, rank(sample)) must hold for the corresponding input arguments (the number of clusters must be less than or equal to the sample size).
The condition ubound(sample, 1) == ubound(center, 1) must hold for the corresponding input arguments.
These conditions are verified only if the library is built with the preprocessor macro CHECK_ENABLED=1.
-
By definition, the number of points in the input
sample must be larger than the specified number of clusters.
-
The
pure procedure(s) documented herein become impure when the ParaMonte library is compiled with preprocessor macro CHECK_ENABLED=1.
By default, these procedures are pure in release build and impure in debug and testing builds. The procedures of this generic interface are always impure when the input rng argument is set to an object of type rngf_type.
- Note
- Dropping the optional arguments can aid runtime performance.
This is particularly relevant when the output of this generic interface is directly passed to the k-means algorithm.
- See also
- setKmeans
setCenter
setMember
setKmeansPP
Arthur, D.; Vassilvitskii, S. (2007). k-means++: the advantages of careful seeding
Example usage ⛓
12 integer(IK) :: ndim, nsam, ncls, itry
13 real(RKG) ,
allocatable :: sample(:,:), center(:,:), disq(:), potential(:)
14 integer(IK) ,
allocatable :: membership(:),
size(:)
15 type(display_type) :: disp
20 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
21 call disp%show(
"! Compute cluster centers based on an input sample and cluster memberships and member-center distances.")
22 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
27 call disp%show(
"ndim = getUnifRand(1, 5); ncls = getUnifRand(1, 5); nsam = getUnifRand(ncls, 2 * ncls);")
31 call disp%show(
"sample = getUnifRand(0., 5., ndim, nsam) ! Create a random sample.")
35 call disp%show(
"call setResized(disq, nsam)")
37 call disp%show(
"call setResized(membership, nsam)")
39 call disp%show(
"call setResized(center, [ndim, ncls])")
41 call disp%show(
"call setResized(potential, ncls)")
43 call disp%show(
"call setResized(size, ncls)")
47 call disp%show(
"call setKmeansPP(rngf, membership, disq, sample, center, size, potential) ! compute the new clusters and memberships.")
48 call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
67 integer(IK) :: funit, i
78 call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
79 open(newunit
= funit, file
= "setKmeansPP.center.txt")
81 write(funit,
"(*(g0,:,','))") i, center(:,i)
84 open(newunit
= funit, file
= "setKmeansPP.sample.txt")
86 write(funit,
"(*(g0,:,','))") membership(i), sample(:,i)
Allocate or resize (shrink or expand) an input allocatable scalar string or array of rank 1....
Generate and return a scalar or a contiguous array of rank 1 of length s1 of randomly uniformly distr...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
This module contains procedures and generic interfaces for resizing allocatable arrays of various typ...
This module contains classes and procedures for computing various statistical quantities related to t...
type(rngf_type) rngf
The scalar constant object of type rngf_type whose presence signified the use of the Fortran intrinsi...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
integer, parameter RKS
The single-precision real kind in Fortran mode. On most platforms, this is an 32-bit real kind.
Generate and return an object of type display_type.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example output ⛓
12+3.09760118,
+4.01347351,
+4.67596292,
+1.24449110,
+4.79837036,
+0.988928974,
+4.86061478,
+1.86515093,
+2.04817796
19call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
21+0.00000000,
+0.00000000,
+0.340963081E-1,
+0.00000000,
+0.387436734E-2,
+0.653119981E-1,
+0.00000000,
+0.334988944E-1,
+0.00000000
23+3,
+4,
+2,
+1,
+2,
+1,
+2,
+5,
+5
25+0.653119981E-1,
+0.379706770E-1,
+0.00000000,
+0.00000000,
+0.334988944E-1
27+1.24449110,
+4.86061478,
+3.09760118,
+4.01347351,
+2.04817796
37+0.515738130,
+3.02103639,
+3.82168722,
+0.714060664E-1
38+2.46166372,
+2.16211796,
+1.28180718,
+1.19970536
45call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
47+0.00000000,
+0.00000000,
+0.00000000,
+1.78996992
51+0.00000000,
+1.78996992,
+0.00000000
53+3.02103639,
+0.515738130,
+3.82168722
54+2.16211796,
+2.46166372,
+1.28180718
64+4.79888678,
+1.00716412
71call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
73+0.00000000,
+14.3771620
89+4.90091419,
+1.72373474,
+2.38117242,
+1.21245646,
+3.18957949,
+3.30897856
90+4.08058310,
+4.79557705,
+4.59610128,
+2.03943491,
+1.72553992,
+4.17742968
97call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
99+2.54363823,
+0.472014874,
+0.00000000,
+4.00754547,
+0.00000000,
+0.00000000
101+1,
+3,
+3,
+2,
+2,
+1
103+2.54363823,
+4.00754547,
+0.472014874
105+3.30897856,
+3.18957949,
+2.38117242
106+4.17742968,
+1.72553992,
+4.59610128
116+1.14988267,
+1.60304523
117+0.300022960E-1,
+3.04963923
118+0.715044141,
+3.05558562
125call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
127+14.8016968,
+0.00000000
145+1.31039524,
+2.36078358,
+4.13939905,
+3.18617964,
+3.27243543,
+2.89390826,
+4.16665030
146+1.68747127,
+4.64989996,
+4.30699682,
+0.235319138E-1,
+2.62257528,
+4.26669216,
+1.83290064
147+4.06491137,
+4.63971186,
+1.69741392,
+1.90310264,
+0.353788435,
+4.00688076,
+4.35267925
154call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
156+0.00000000,
+0.831545353,
+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000,
+7.66279030
158+4,
+5,
+1,
+2,
+3,
+5,
+5
160+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000,
+8.49433517
162+4.13939905,
+3.18617964,
+3.27243543,
+1.31039524,
+2.89390826
163+4.30699682,
+0.235319138E-1,
+2.62257528,
+1.68747127,
+4.26669216
164+1.69741392,
+1.90310264,
+0.353788435,
+4.06491137,
+4.00688076
174+3.39266276,
+2.42241621,
+2.80469370,
+0.854519904,
+4.38353205
175+0.620661080,
+1.61410904,
+0.478409827,
+0.732208192,
+1.41081989
176+0.778235793,
+0.114700198E-1,
+2.78694415,
+2.43016815,
+1.54042244
177+3.66197371,
+3.86894178,
+0.353656113,
+4.93827820,
+1.48715794
178+1.39233327,
+1.44989729,
+4.34943295,
+4.95386887,
+4.16783524
185call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
187+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000
191+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000
193+3.39266276,
+0.854519904,
+4.38353205,
+2.80469370,
+2.42241621
194+0.620661080,
+0.732208192,
+1.41081989,
+0.478409827,
+1.61410904
195+0.778235793,
+2.43016815,
+1.54042244,
+2.78694415,
+0.114700198E-1
196+3.66197371,
+4.93827820,
+1.48715794,
+0.353656113,
+3.86894178
197+1.39233327,
+4.95386887,
+4.16783524,
+4.34943295,
+1.44989729
207+2.78194094,
+2.07066584,
+2.54888511,
+1.01118147,
+1.41291142
208+0.315136909,
+3.63018084,
+3.14631414,
+4.36529112,
+4.01214123
215call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
217+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000
221+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000,
+0.00000000
223+2.54888511,
+2.78194094,
+1.01118147,
+1.41291142,
+2.07066584
224+3.14631414,
+0.315136909,
+4.36529112,
+4.01214123,
+3.63018084
234+0.713005364,
+0.361892879,
+0.748941004,
+0.368515253
235+3.42950010,
+1.48842669,
+4.12447643,
+4.43461227
242call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
244+0.00000000,
+0.00000000,
+0.240907997,
+0.00000000
248+0.00000000,
+0.240907997,
+0.00000000
250+0.713005364,
+0.368515253,
+0.361892879
251+3.42950010,
+4.43461227,
+1.48842669
261+3.23012495,
+0.428308249,
+3.80425668,
+4.68496466,
+0.673015118,
+3.38467789,
+4.53701115,
+0.359097123E-1,
+2.58815289,
+0.239347816
268call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
270+0.238866098E-1,
+0.357060470E-1,
+0.00000000,
+0.218902417E-1,
+0.00000000,
+0.00000000,
+0.00000000,
+0.413870625E-1,
+0.634452105,
+0.00000000
272+1,
+2,
+4,
+3,
+5,
+1,
+3,
+2,
+1,
+2
274+0.658338726,
+0.770931095E-1,
+0.218902417E-1,
+0.00000000,
+0.00000000
276+3.38467789,
+0.239347816,
+4.53701115,
+3.80425668,
+0.673015118
Postprocessing of the example output ⛓
3import matplotlib.pyplot
as plt
11fig = plt.figure(figsize = 1.25 * np.array([6.4, 4.8]), dpi = 200)
14parent = os.path.basename(os.path.dirname(__file__))
15pattern = parent +
"*.txt"
17fileList = glob.glob(pattern)
22 kind = file.split(
".")[1]
23 prefix = file.split(
".")[0]
24 df = pd.read_csv(file, delimiter =
",", header =
None)
27 ax.scatter ( df.values[:,1]
34 legends.append(
"center")
35 elif kind ==
"sample":
36 ax.scatter ( df.values[:,1]
41 legends.append(
"sample")
43 sys.exit(
"Ambiguous file exists: {}".format(file))
45 ax.legend(legends, fontsize = fontsize)
46 plt.xticks(fontsize = fontsize - 2)
47 plt.yticks(fontsize = fontsize - 2)
48 ax.set_xlabel(
"X", fontsize = 17)
49 ax.set_ylabel(
"Y", fontsize = 17)
50 ax.set_title(
"Membership Scatter Plot", fontsize = fontsize)
53 plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
54 ax.tick_params(axis =
"y", which =
"minor")
55 ax.tick_params(axis =
"x", which =
"minor")
56 ax.set_axisbelow(
True)
59 plt.savefig(prefix +
".png")
61 sys.exit(
"Ambiguous file list exists.")
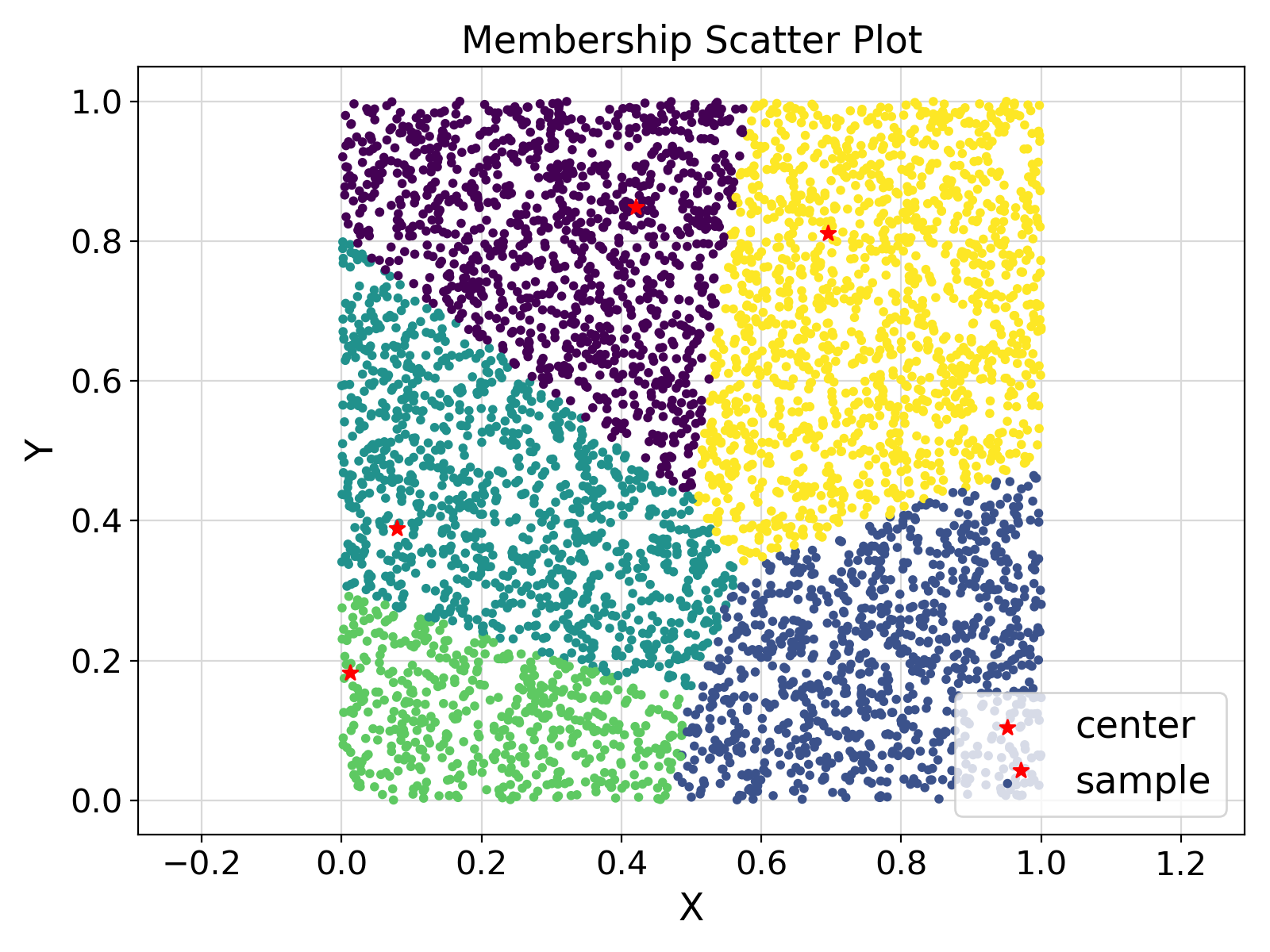
Visualization of the example output ⛓
- Test:
- test_pm_clustering
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab If you use or redistribute ideas based on this generic interface implementation, you should also cite the original k-means++ article:
Arthur, D.; Vassilvitskii, S. (2007). k-means++: the advantages of careful seeding
- Author:
- Amir Shahmoradi, September 1, 2012, 12:00 AM, National Institute for Fusion Studies, The University of Texas at Austin
Definition at line 787 of file pm_clustering.F90.