Compute and return an iteratively-refined set of cluster centers given the input sample using the k-means approach.
More...
Compute and return an iteratively-refined set of cluster centers given the input sample using the k-means approach.
The k-means clustering is a method of vector quantization, originally from signal processing, that aims to partition n observations into \(k\) clusters in which each observation belongs to the cluster with the nearest mean (cluster centers or cluster centroid), serving as a prototype of the cluster.
This results in a partitioning of the data space into Voronoi cells.
The k-means clustering minimizes within-cluster variances (squared Euclidean distances), but not regular Euclidean distances, which would be the more difficult Weber problem:
the mean optimizes squared errors, whereas only the geometric median minimizes Euclidean distances.
For instance, better Euclidean solutions can be found using k-medians and k-medoids.
The problem is computationally difficult (NP-hard); however, efficient heuristic algorithms converge quickly to a local optimum.
These are usually similar to the expectation-maximization algorithm for mixtures of Gaussian distributions via an iterative refinement approach employed by both k-means and Gaussian mixture modeling.
They both use cluster centers to model the data; however, k-means clustering tends to find clusters of comparable spatial extent, while the Gaussian mixture model allows clusters to have different shapes.
Algorithm
Given a set of observations \((x_1, x_2, \ldots, x_n)\), where each observation is a \(d\)-dimensional real vector, the k-means clustering aims to partition the \(n\) observations into \(k\) ( \(\leq n\)) sets \(S = \{S_1, S_2, \ldots, S_k\}\) so as to minimize the within-cluster sum of squares (WCSS) (i.e. variance).
Formally, the objective is to find:
\begin{equation}
\underset{\mathbf{S}}{\up{arg\,min}}
\sum_{i=1}^{k} \sum_{\mathbf{x} \in S_{i}} \left\|\mathbf{x} -{\boldsymbol{\mu}}_{i}\right\|^{2}
= {\underset{\mathbf{S}}{\up{arg\,min}}}\sum_{i=1}^{k}|S_{i}|\up{Var}S_{i} ~,
\end{equation}
where \(\mu_i\) is the mean (also called centroid) of points in \(S_{i}\), i.e.
\begin{equation}
{\boldsymbol {\mu_{i}}} = {\frac{1}{|S_{i}|}} \sum_{\mathbf{x} \in S_{i}} \mathbf{x} ~,
\end{equation}
where \(|S_{i}|\) is the size of \(S_{i}\), and \(\|\cdot\|\) is the \(L^2\)-norm.
This is equivalent to minimizing the pairwise squared deviations of points in the same cluster:
\begin{equation}
\underset{\mathbf{S}}{\up{arg\,min}} \sum_{i=1}^{k}\,{\frac {1}{|S_{i}|}}\,\sum_{\mathbf{x}, \mathbf{y} \in S_{i}}\left\|\mathbf{x} - \mathbf{y} \right\|^{2} ~,
\end{equation}
The equivalence can be deduced from identity
\begin{equation}
|S_{i}|\sum_{\mathbf{x} \in S_{i}}\left\|\mathbf{x} -{\boldsymbol{\mu}}_{i}\right\|^{2} =
{\frac{1}{2}}\sum _{\mathbf {x} ,\mathbf {y} \in S_{i}}\left\|\mathbf {x} -\mathbf {y} \right\|^{2} ~.
\end{equation}
Since the total variance is constant, this is equivalent to maximizing the sum of squared deviations between points in different clusters.
This deterministic relationship is also related to the law of total variance in probability theory.
Performance improvements
The k-means++ seeding method yields considerable improvement in the final error of k-means algorithm.
For more information, see the documentation of setKmeansPP.
- Note
- The metric used within this generic interface is the Euclidean distance.
- Parameters
-
| [in,out] | rng | : The input/output scalar that can be an object of,
-
type rngf_type, implying the use of intrinsic Fortran uniform RNG.
-
type xoshiro256ssw_type, implying the use of xoshiro256** uniform RNG.
(optional, If this argument is present, then all intent(inout) arguments below have intent(out) argument and will be initialized using the k-means++ algorithm.
If this argument is missing, then the user must initialize all of the following arguments with intent(inout) via k-means++ or any other method before passing them to this generic interface.) |
| [in,out] | membership | : The input/output vector of shape (1:nsam) of type integer of default kind IK, containing the membership of each input sample in sample from its nearest cluster center, such that cluster(membership(i)) is the nearest cluster center to the ith sample sample(:, i) at a squared-distance of disq(i).
If the argument rng is missing, then membership has intent(inout) and must be properly initialized before calling this routine.
If the argument rng is present, then membership has intent(out) and will contain the final cluster memberships on output.
|
| [in,out] | disq | : The input/output vector of shape (1:nsam) of the same type and kind as the input argument sample, containing the Euclidean squared distance of each input sample in sample from its nearest cluster center.
If the argument rng is missing, then disq has intent(inout) and must be properly initialized before calling this routine.
If the argument rng is present, then disq has intent(out) and will contain the final squared-distances from cluster centers on output.
|
| [in] | sample | : The input vector, or matrix of,
-
type
real of kind any supported by the processor (e.g., RK, RK32, RK64, or RK128),
containing the sample of nsam points in a ndim-dimensional space whose corresponding cluster centers must be computed.
-
If
sample is a vector of shape (1 : nsam) and center is a vector of shape (1 : ncls), then the input sample must be a collection of nsam points (in univariate space).
-
If
sample is a matrix of shape (1 : ndim, 1 : nsam) and center is a matrix of shape (1 : ndim, 1 : ncls), then the input sample must be a collection of nsam points (in ndim-dimensional space).
|
| [in,out] | center | : The input/output vector of shape (1:ncls) or matrix of shape (1 : ndim, 1 : ncls) of the same type and kind as the input argument sample, containing the set of ncls unique cluster centers (centroids) computed based on the input sample memberships and minimum distances.
If the argument rng is missing, then center has intent(inout) and must be properly initialized before calling this routine.
If the argument rng is present, then center has intent(out) and will contain the final cluster centers on output.
|
| [in,out] | size | : The input/output vector of shape (1:ncls) type integer of default kind IK, containing the sizes (number of members) of the clusters with the corresponding centers output in the argument center.
If the argument rng is missing, then size has intent(inout) and must be properly initialized before calling this routine.
If the argument rng is present, then size has intent(out) and will contain the final cluster sizes (member counts) on output.
|
| [in,out] | potential | : The input/output vector of shape (1:ncls) of the same type and kind as the input argument sample, the ith element of which contains the sum of squared distances of all members of the ith cluster from the cluster center as output in the ith element of center.
If the argument rng is missing, then potential has intent(inout) and must be properly initialized before calling this routine (although its values are not explicitly referenced).
If the argument rng is present, then potential has intent(out) and will contain the final cluster potentials (sums of squared distances from cluster centers) on output.
|
| [out] | failed | : The output scalar of type logical of default kind LK that is .true. if and only if the algorithm fails to converge within the user-specified or default criteria for convergence.
Failure occurs only if any(size < minsize) .or. maxniter < niter.
|
| [out] | niter | : The output scalar of type integer of default kind IK, containing the number of refinement iterations performed within the algorithm to achieve convergence.
An output niter value larger than the input maxniter implies lack of convergence before return.
(optional. If missing, the number of refinement iterations will not be output.) |
| [in] | maxniter | : The input non-negative scalar of type integer of default kind IK, containing the maximum number of refinement iterations allowed within the algorithm to achieve convergence.
If convergence does not occur within the maximum specified value, the output arguments can be passed again as is to the generic interface (without the optional rng argument) to continue the refinement iterations until convergence.
A reasonable choice can be 300 or comparable values.
(optional, default = 300.) |
| [in] | minsize | : The input non-negative scalar of type integer of default kind IK, containing the minimum allowed size of each cluster.
If any cluster has any number of members below the specified minsize, the algorithm will return without achieving convergence.
The situation can be detected of any element of the output size is smaller than the specified minsize.
A reasonable choice can be ndim = ubound(sample, 1) or comparable values although any non-negative value including zero is possible.
(optional, default = 1.) |
| [in] | nfail | : The input non-negative scalar of type integer of default kind IK, containing the number of times the k-means algorithm is allowed to fail before returning without convergence.
(optional. It can be present only if the argument rng is also present, allowing random initializations in case of failures.) |
Possible calling interfaces ⛓
call setKmeansPP(membership(
1 : nsam) , disq(
1 : nsam), sample(
1 : nsam) , center(
1 : ncls) ,
size(
1 : ncls), potential(
1 : ncls), failed, niter
= niter, maxniter
= maxniter, minsize
= minsize)
call setKmeansPP(membership(
1 : nsam) , disq(
1 : nsam), sample(
1 : ndim,
1 : nsam), center(
1 : ndim,
1 : ncls),
size(
1 : ncls), potential(
1 : ncls), failed, niter
= niter, maxniter
= maxniter, minsize
= minsize)
call setKmeansPP(rng, membership(
1 : nsam) , disq(
1 : nsam), sample(
1 : nsam) , center(
1 : ncls) ,
size(
1 : ncls), potential(
1 : ncls), failed, niter
= niter, maxniter
= maxniter, minsize
= minsize, nfail
= nfail)
call setKmeansPP(rng, membership(
1 : nsam) , disq(
1 : nsam), sample(
1 : ndim,
1 : nsam), center(
1 : ndim,
1 : ncls),
size(
1 : ncls), potential(
1 : ncls), failed, niter
= niter, maxniter
= maxniter, minsize
= minsize, nfail
= nfail)
Compute and return an asymptotically optimal set of cluster centers for the input sample,...
Compute and return an iteratively-refined set of cluster centers given the input sample using the k-m...
This module contains procedures and routines for the computing the Kmeans clustering of a given set o...
- Warning
- If the argument
rng is present, then all arguments associated with setKmeansPP equally apply to this generic interface.
The condition ubound(center, rank(center)) > 0 must hold for the corresponding input arguments.
The condition ubound(sample, rank(sample)) == ubound(disq, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) == ubound(size, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) == ubound(potential, 1) must hold for the corresponding input arguments.
The condition ubound(sample, rank(sample)) == ubound(membership, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) <= ubound(sample, rank(sample)) must hold for the corresponding input arguments (the number of clusters must be less than or equal to the sample size).
The condition ubound(sample, 1) == ubound(center, 1) must hold for the corresponding input arguments.
The condition 0 <= maxniter must hold for the corresponding input arguments.
The condition 0 <= minsize must hold for the corresponding input arguments.
The condition 0 <= nfail must hold for the corresponding input arguments.
These conditions are verified only if the library is built with the preprocessor macro CHECK_ENABLED=1.
-
The
pure procedure(s) documented herein become impure when the ParaMonte library is compiled with preprocessor macro CHECK_ENABLED=1.
By default, these procedures are pure in release build and impure in debug and testing builds. The procedures of this generic interface are always impure when the input rng argument is set to an object of type rngf_type.
- See also
- setKmeans
setCenter
setMember
setKmeansPP
k-means clustering
Example usage ⛓
14 integer(IK) :: ndim, nsam, ncls, itry, niter
15 real(RKG) ,
allocatable :: sample(:,:), center(:,:), disq(:), potential(:)
16 integer(IK) ,
allocatable :: membership(:),
size(:)
17 type(display_type) :: disp
22 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
23 call disp%show(
"! Compute cluster centers based on an input sample and cluster memberships and member-center distances.")
24 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
29 call disp%show(
"ndim = getUnifRand(1, 5); ncls = getUnifRand(1, 5); nsam = getUnifRand(ncls, 2 * ncls);")
33 call disp%show(
"sample = getUnifRand(0., 5., ndim, nsam) ! Create a random sample.")
37 call disp%show(
"call setResized(disq, nsam)")
39 call disp%show(
"call setResized(membership, nsam)")
41 call disp%show(
"call setResized(center, [ndim, ncls])")
43 call disp%show(
"call setResized(potential, ncls)")
45 call disp%show(
"call setResized(size, ncls)")
49 call disp%show(
"call setKmeans(rngf, membership, disq, sample, center, size, potential, failed, niter) ! compute the new clusters and memberships.")
50 call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
73 integer(IK) :: funit, i
84 call setKmeansPP(
rngf, membership, disq, sample, center, size, potential)
85 open(newunit
= funit, file
= "setKmeansPP.center.txt")
87 write(funit,
"(*(g0,:,','))") i, center(:,i)
90 open(newunit
= funit, file
= "setKmeansPP.sample.txt")
92 write(funit,
"(*(g0,:,','))") membership(i), sample(:,i)
95 call setKmeans(membership, disq, sample, center, size, potential, failed)
96 open(newunit
= funit, file
= "setKmeans.center.txt")
98 write(funit,
"(*(g0,:,','))") i, center(:,i)
101 open(newunit
= funit, file
= "setKmeans.sample.txt")
103 write(funit,
"(*(g0,:,','))") membership(i), sample(:,i)
Allocate or resize (shrink or expand) an input allocatable scalar string or array of rank 1....
Generate and return a scalar or a contiguous array of rank 1 of length s1 of randomly uniformly distr...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
This module contains procedures and generic interfaces for resizing allocatable arrays of various typ...
This module contains classes and procedures for computing various statistical quantities related to t...
type(rngf_type) rngf
The scalar constant object of type rngf_type whose presence signified the use of the Fortran intrinsi...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
integer, parameter RKS
The single-precision real kind in Fortran mode. On most platforms, this is an 32-bit real kind.
Generate and return an object of type display_type.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example output ⛓
12+3.09051657,
+1.26056886
13+2.13868189,
+2.20415926
14+0.165876448,
+4.62417698
15+3.33872294,
+4.93674469
16+4.05802822,
+0.188024044
23call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
29+0.00000000,
+0.00000000
33+0.00000000,
+0.00000000
35+1.26056886,
+3.09051657
36+2.20415926,
+2.13868189
37+4.62417698,
+0.165876448
38+4.93674469,
+3.33872294
39+0.188024044,
+4.05802822
49+0.964425206,
+2.01776719,
+0.694973171,
+2.03159618
50+2.78230238,
+4.44708538,
+2.54506230,
+3.24920464
51+2.11412859,
+4.58920431,
+3.20349455,
+2.51365948
52+0.577360690,
+2.93278432,
+3.28652120,
+3.56598997
59call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
65+0.00000000,
+2.69745898,
+1.59492815,
+1.17197776
69+0.00000000,
+8.98029804
71+0.964425206,
+1.58144557
72+2.78230238,
+3.41378403
73+2.11412859,
+3.43545294
74+0.577360690,
+3.26176524
84+3.49306345,
+4.84981823,
+0.482063591,
+1.34671211
85+2.88380194,
+1.71131730,
+1.93086922,
+0.324480832
86+2.77829385,
+0.889991820,
+0.842357278,
+0.735339522E-1
93call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
99+1.69529724,
+1.69529688,
+0.979797423,
+0.979797602
103+6.78118849,
+3.91918969
105+4.17144108,
+0.914387822
106+2.29755974,
+1.12767506
107+1.83414280,
+0.457945615
117+2.67365503,
+1.73565805,
+1.08126378
118+0.780725479,
+3.58734083,
+2.24011850
119+3.19889784,
+4.77777624,
+4.48111963
120+0.197524428,
+4.13293648,
+1.16237903
121+1.01525640,
+2.67101073,
+1.36856318
128call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
134+0.00000000,
+3.21295643,
+3.21295691
138+12.8518257,
+0.00000000
140+1.40846086,
+2.67365503
141+2.91372967,
+0.780725479
142+4.62944794,
+3.19889784
143+2.64765787,
+0.197524428
144+2.01978683,
+1.01525640
154+3.36679554,
+2.60843635,
+3.53108978,
+0.765241981,
+4.52701473,
+1.37547374,
+2.27166533
155+0.216679573,
+2.14455223,
+0.186871588,
+4.49863815,
+0.525346994,
+3.02717018,
+4.27078485
162call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
168+0.203566492,
+0.00000000,
+0.919158086E-1,
+0.580307066,
+0.563083470,
+0.00000000,
+0.580307126
170+2,
+4,
+2,
+1,
+2,
+3,
+1
172+2.32122850,
+1.13431323,
+0.00000000,
+0.00000000
174+1.51845360,
+3.80830002,
+1.37547374,
+2.60843635
175+4.38471127,
+0.309632719,
+3.02717018,
+2.14455223
185+3.42633343,
+4.87914419,
+4.52042580
186+0.704374611,
+3.36541390,
+2.75196648
193call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
199+0.00000000,
+0.00000000,
+0.00000000
203+0.00000000,
+0.00000000,
+0.00000000
205+4.52042580,
+3.42633343,
+4.87914419
206+2.75196648,
+0.704374611,
+3.36541390
216+0.335005224,
+3.19071317,
+1.81889749,
+4.55224800,
+2.48839808,
+3.59679031,
+1.09610105
217+1.55076122,
+2.90543056,
+1.28109694,
+1.30711412,
+3.08899903,
+0.990077555,
+4.96392870
218+0.603176951,
+2.59593105,
+2.10227966,
+4.72403002,
+1.45300925,
+4.23028374,
+3.30410004
219+3.65917563,
+4.09179115,
+3.23305631,
+1.01636267,
+1.72277927,
+4.31291151,
+4.91639090
220+0.135381818,
+2.18604064,
+1.21946192,
+0.665618479,
+0.938138664,
+1.07311583,
+2.86370158
227call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
233+0.00000000,
+1.07133758,
+0.00000000,
+4.67686510,
+4.67686462,
+6.47388220,
+7.29835606
235+3,
+2,
+4,
+1,
+1,
+2,
+2
237+18.7074585,
+18.0575886,
+0.00000000,
+0.00000000
239+3.52032304,
+2.62786818,
+0.335005224,
+1.81889749
240+2.19805670,
+2.95314574,
+1.55076122,
+1.28109694
241+3.08851957,
+3.37677169,
+0.603176951,
+2.10227966
242+1.36957097,
+4.44036484,
+3.65917563,
+3.23305631
243+0.801878572,
+2.04095268,
+0.135381818,
+1.21946192
253+2.67327094,
+4.58450317,
+2.33191395
254+4.88044834,
+3.23485970,
+4.64746809
255+4.03332853,
+2.09349895,
+3.93679595
256+0.102661848,
+1.45601845,
+4.11710501
257+3.25221419,
+2.74718666,
+0.947423279
264call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
270+3.05258465,
+3.05258369,
+0.00000000
274+0.00000000,
+12.2103367
276+2.33191395,
+3.62888718
277+4.64746809,
+4.05765390
278+3.93679595,
+3.06341362
279+4.11710501,
+0.779340148
280+0.947423279,
+2.99970055
290+4.38341188,
+3.85437036
297call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
303+0.699711740E-1,
+0.699713007E-1
319+4.91269827,
+3.94869113,
+3.45396805,
+0.466822684,
+2.31161690,
+3.39903259,
+2.29473591
320+4.38696861,
+3.01287937,
+3.21296883,
+0.671095252,
+1.87571108,
+2.92932224,
+2.89692926
327call setKmeans(
rngf, membership, disq, sample, center, size, potential, failed, niter)
333+0.00000000,
+0.122701362,
+0.474904366E-1,
+0.00000000,
+0.260792822,
+0.555969737E-1,
+0.260792941
335+1,
+4,
+4,
+2,
+3,
+4,
+3
337+0.00000000,
+0.00000000,
+1.04317153,
+0.593893051
339+4.91269827,
+0.466822684,
+2.30317640,
+3.60056400
340+4.38696861,
+0.671095252,
+2.38632011,
+3.05172348
Postprocessing of the example output ⛓
3import matplotlib.pyplot
as plt
12prefixes = {
"setKmeansPP" :
"Memberships based on k-means++."
13 ,
"setKmeans" :
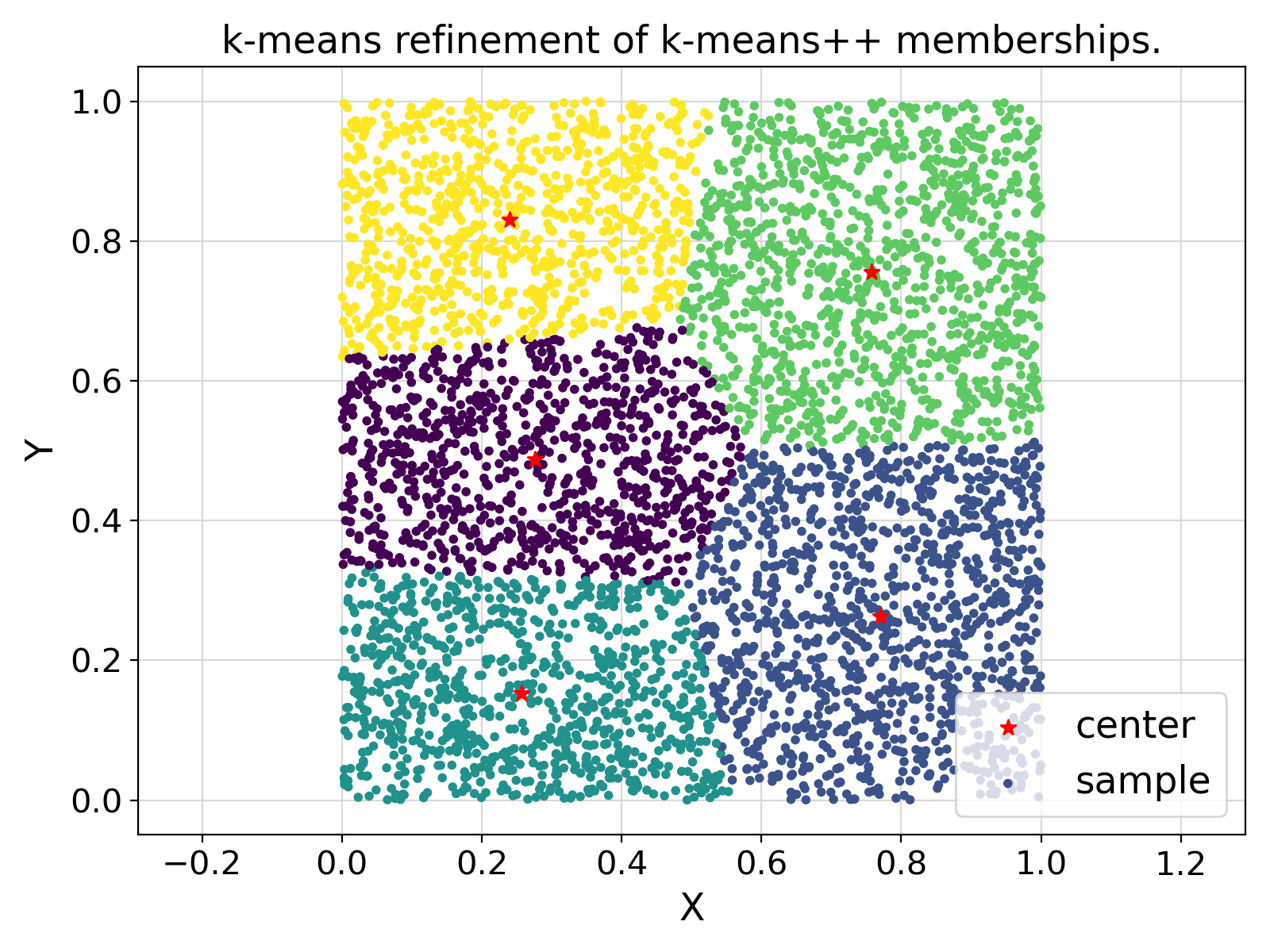
"k-means refinement of k-means++ memberships."
15for prefix
in list(prefixes.keys()):
18 fig = plt.figure(figsize = 1.25 * np.array([6.4, 4.8]), dpi = 200)
21 fileList = glob.glob(prefix +
".*.txt")
22 if len(fileList) == 2:
26 kind = file.split(
".")[1]
27 df = pd.read_csv(file, delimiter =
",", header =
None)
30 ax.scatter ( df.values[:,1]
37 legends.append(
"center")
38 elif kind ==
"sample":
39 ax.scatter ( df.values[:,1]
44 legends.append(
"sample")
46 sys.exit(
"Ambiguous file exists: {}".format(file))
48 ax.legend(legends, fontsize = fontsize)
49 plt.xticks(fontsize = fontsize - 2)
50 plt.yticks(fontsize = fontsize - 2)
51 ax.set_xlabel(
"X", fontsize = 17)
52 ax.set_ylabel(
"Y", fontsize = 17)
53 ax.set_title(prefixes[prefix], fontsize = fontsize)
56 plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
57 ax.tick_params(axis =
"y", which =
"minor")
58 ax.tick_params(axis =
"x", which =
"minor")
59 ax.set_axisbelow(
True)
62 plt.savefig(prefix +
".png")
66 sys.exit(
"Ambiguous file list exists.")
Visualization of the example output ⛓
- Test:
- test_pm_clustering
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, September 1, 2012, 12:00 AM, National Institute for Fusion Studies, The University of Texas at Austin
Definition at line 1292 of file pm_clustering.F90.