Compute and return the centers of the clusters corresponding to the input sample, cluster membership IDs, and sample distances-squared from their corresponding cluster centers.
This generic interface is the second step in the iterative process of refining a set of initial cluster centers toward a final set of clusters and their members.
As such, this generic interface is of little use with invoking setMember beforehand.
The metric used within this generic interface is the Euclidean distance.
- Parameters
-
| [in] | membership | : The input vector of shape (1:nsam) of type integer of default kind IK, containing the membership of each input sample in sample from its nearest cluster center, such that cluster(membership(i)) is the nearest cluster center to the ith sample sample(:, i) at a squared-distance of disq(i).
|
| [in] | disq | : The input vector of shape (1:nsam) of the same type and kind as the input argument sample, containing the Euclidean squared distance of each input sample in sample from its nearest cluster center.
|
| [in] | sample | : The input vector, or matrix of,
-
type
real of kind any supported by the processor (e.g., RK, RK32, RK64, or RK128),
containing the sample of nsam points in a ndim-dimensional space whose corresponding cluster centers must be computed.
-
If
sample is a vector of shape (1 : nsam) and center is a vector of shape (1 : ncls), then the input sample must be a collection of nsam points (in univariate space).
-
If
sample is a matrix of shape (1 : ndim, 1 : nsam) and center is a matrix of shape (1 : ndim, 1 : ncls), then the input sample must be a collection of nsam points (in ndim-dimensional space).
|
| [out] | center | : The output vector of shape (1:ncls) or matrix of shape (1 : ndim, 1 : ncls) of the same type and kind as the input argument sample, containing the set of ncls cluster centers (centroids) computed based on the input sample memberships and minimum distances.
|
| [out] | size | : The output vector of shape (1:ncls) type integer of default kind IK, containing the sizes (number of members) of the clusters with the corresponding centers output in the argument center.
|
| [out] | potential | : The output vector of shape (1:ncls) of the same type and kind as the input argument sample, the ith element of which contains the sum of squared distances of all members of the ith cluster from the cluster center as output in the ith element of center.
|
Possible calling interfaces ⛓
call setCenter(sample(
1 : nsam) , membership(
1 : nsam) , disq(
1 : nsam), center(
1 : ncls) ,
size(
1 : ncls), potential(
1 : ncls))
call setCenter(sample(
1 : ndim,
1 : nsam) , membership(
1 : nsam) , disq(
1 : nsam), center(
1 : ndim,
1 : ncls),
size(
1 : ncls), potential(
1 : ncls))
Compute and return the centers of the clusters corresponding to the input sample, cluster membership ...
This module contains procedures and routines for the computing the Kmeans clustering of a given set o...
- Warning
- The condition
ubound(center, rank(center)) > 0 must hold for the corresponding input arguments.
The condition ubound(sample, rank(sample)) == ubound(disq, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) == ubound(size, 1) must hold for the corresponding input arguments.
The condition ubound(center, rank(center)) == ubound(potential, 1) must hold for the corresponding input arguments.
The condition ubound(sample, rank(sample)) == ubound(membership, 1) must hold for the corresponding input arguments.
The condition ubound(sample, 1) == ubound(center, 1) .or. (rank(sample) == 1 .and. rank(center) == 1) must hold for the corresponding input arguments.
The condition all(0 < membership .and. membership <= ubound(center, rank(center))) must hold for the corresponding input arguments.
The condition all(0 <= disq) must hold for the corresponding input arguments.
These conditions are verified only if the library is built with the preprocessor macro CHECK_ENABLED=1.
-
The
pure procedure(s) documented herein become impure when the ParaMonte library is compiled with preprocessor macro CHECK_ENABLED=1.
By default, these procedures are pure in release build and impure in debug and testing builds.
- See also
- setKmeans
setCenter
setMember
setKmeansPP
Example usage ⛓
13 integer(IK) :: ndim, nsam, ncls
14 real(RKG) ,
allocatable :: sample(:,:), center(:,:), disq(:), potential(:)
15 integer(IK) ,
allocatable :: membership(:),
size(:)
16 type(display_type) :: disp
21 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
22 call disp%show(
"! Compute cluster centers based on an input sample and cluster memberships and member-center distances.")
23 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
27 call disp%show(
"ndim = getUnifRand(1, 5); nsam = getUnifRand(1, 5); ncls = getUnifRand(1, 5);")
31 call disp%show(
"center = getUnifRand(0., 5., ndim, ncls) ! initialize random centers.")
35 call disp%show(
"sample = getUnifRand(0., 5., ndim, nsam) ! Create a random sample.")
39 call disp%show(
"call setResized(disq, nsam)")
41 call disp%show(
"call setResized(membership, nsam)")
43 call disp%show(
"call setResized(potential, ncls)")
45 call disp%show(
"call setResized(size, ncls)")
47 call disp%show(
"call setMember(membership, disq, sample, center) ! get sample points memberships.")
48 call setMember(membership, disq, sample, center)
51 call disp%show(
"call setCenter(membership, disq, sample, center, size, potential) ! now compute the new clusters.")
52 call setCenter(membership, disq, sample, center, size, potential)
61 call disp%show(
"call setCenter(membership, disq, sample(1,:), center(1,:), size, potential) ! sample points memberships in one-dimension.")
62 call setCenter(membership, disq, sample(
1,:), center(
1,:), size, potential)
76 integer(IK) :: funit, i
87 call setMember(membership, disq, sample, center)
88 open(newunit
= funit, file
= "setMember.center.txt")
90 write(funit,
"(*(g0,:,','))") i, center(:,i)
93 open(newunit
= funit, file
= "setMember.sample.txt")
95 write(funit,
"(*(g0,:,','))") membership(i), sample(:,i)
98 call setCenter(membership, disq, sample, center, size, potential)
99 open(newunit
= funit, file
= "setCenter.center.txt")
101 write(funit,
"(*(g0,:,','))") i, center(:,i)
104 open(newunit
= funit, file
= "setCenter.sample.txt")
106 write(funit,
"(*(g0,:,','))") membership(i), sample(:,i)
Allocate or resize (shrink or expand) an input allocatable scalar string or array of rank 1....
Compute and return the memberships and minimum distances of a set of input points with respect to the...
Generate and return a scalar or a contiguous array of rank 1 of length s1 of randomly uniformly distr...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
This module contains procedures and generic interfaces for resizing allocatable arrays of various typ...
This module contains classes and procedures for computing various statistical quantities related to t...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
integer, parameter RKS
The single-precision real kind in Fortran mode. On most platforms, this is an 32-bit real kind.
Generate and return an object of type display_type.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example output ⛓
15+3.64694548,
+1.80137849,
+2.11540985
20call setMember(membership, disq, sample, center)
22call setCenter(membership, disq, sample, center, size, potential)
30call setCenter(membership, disq, sample(
1,:), center(
1,:), size, potential)
Postprocessing of the example output ⛓
3import matplotlib.pyplot
as plt
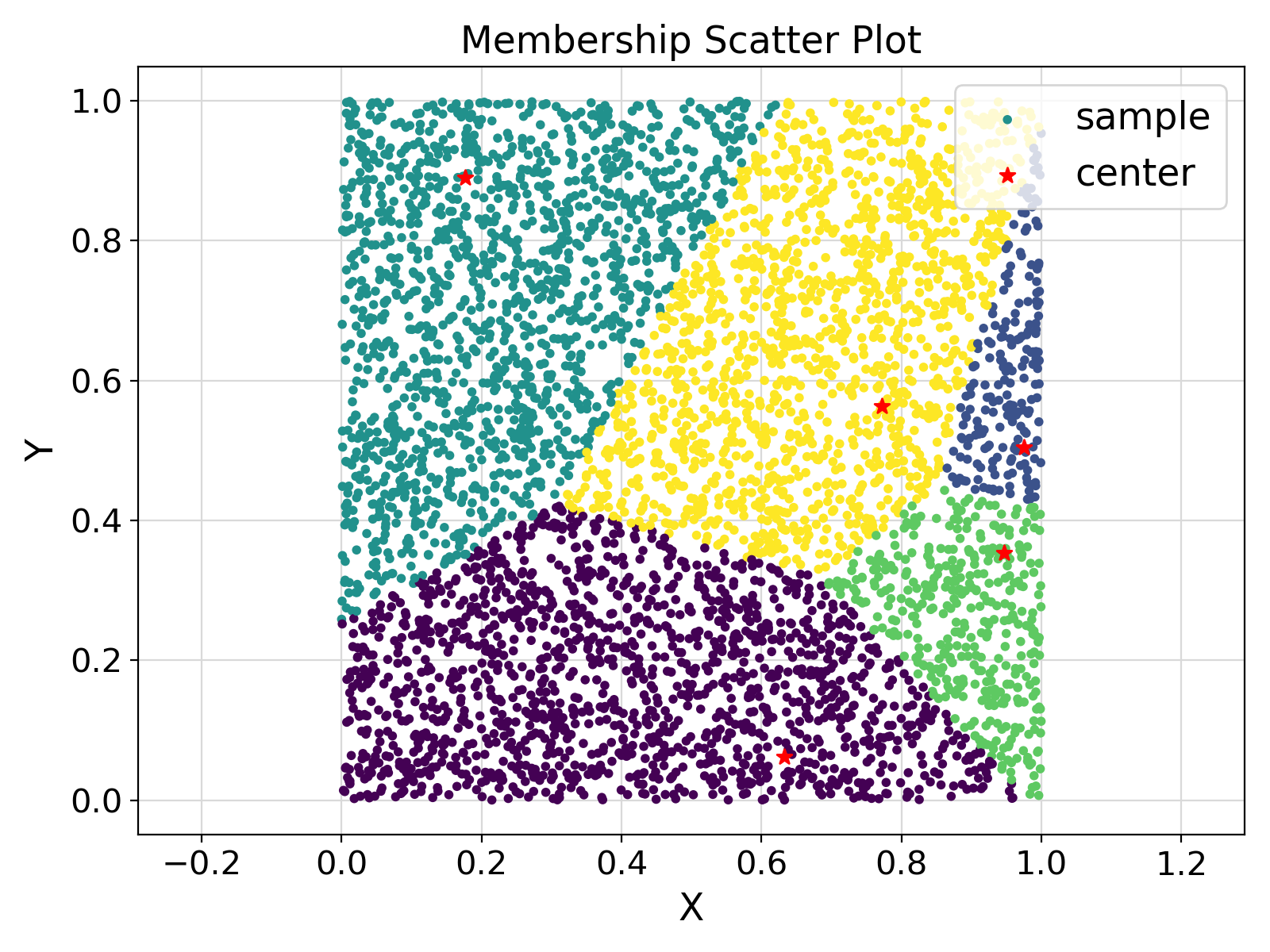
12prefixes = {
"setMember" :
"Memberships based on initial centers."
13 ,
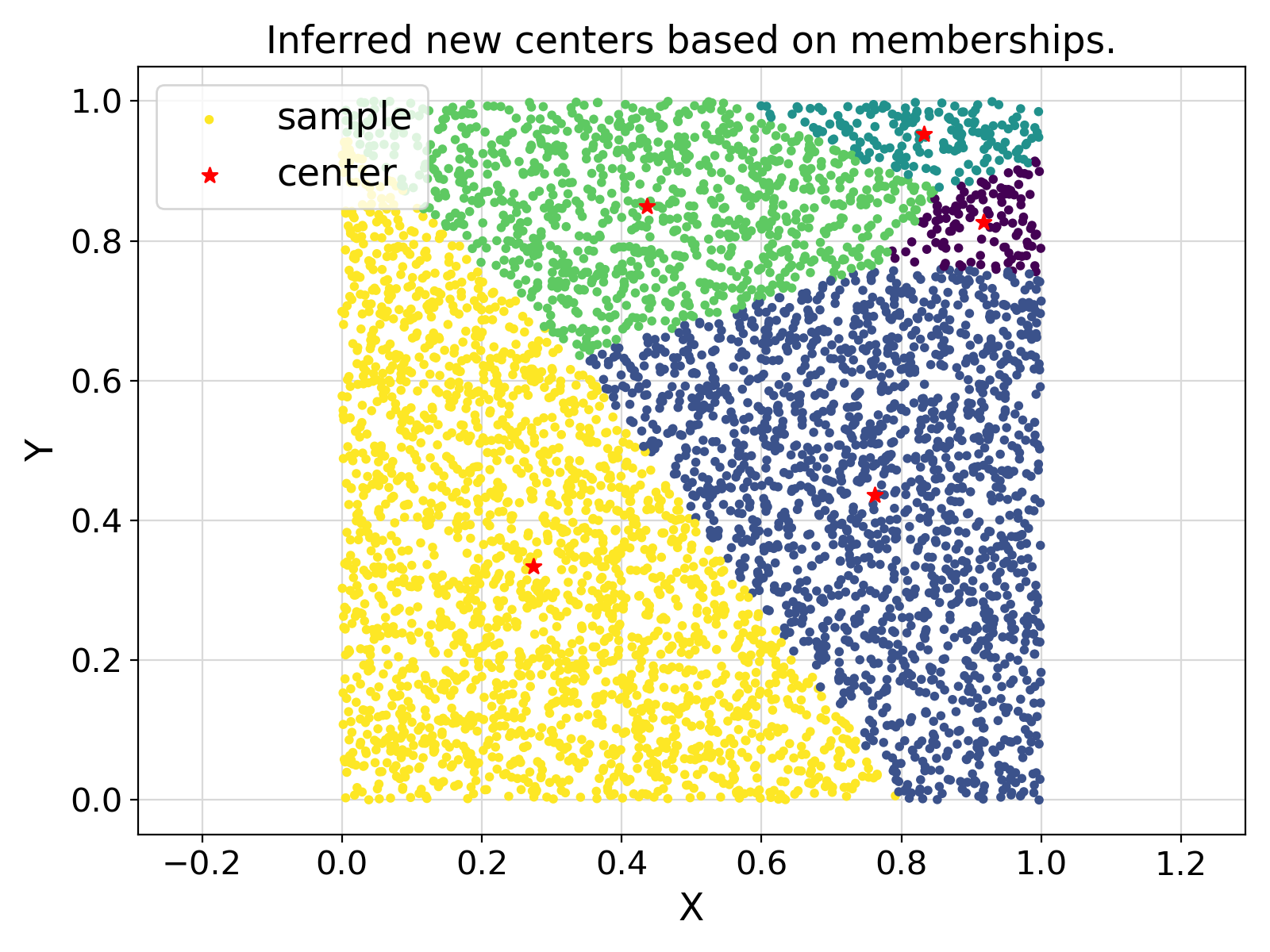
"setCenter" :
"Inferred new centers based on memberships."
15for prefix
in list(prefixes.keys()):
18 fig = plt.figure(figsize = 1.25 * np.array([6.4, 4.8]), dpi = 200)
21 fileList = glob.glob(prefix +
"*.txt")
22 if len(fileList) == 2:
25 kind = file.split(
".")[1]
26 df = pd.read_csv(file, delimiter =
",", header =
None)
29 ax.scatter ( df.values[:, 1]
36 legends.append(
"center")
37 elif kind ==
"sample":
38 ax.scatter ( df.values[:, 1]
43 legends.append(
"sample")
45 sys.exit(
"Ambiguous file exists: {}".format(file))
47 ax.legend(legends, fontsize = fontsize)
48 plt.xticks(fontsize = fontsize - 2)
49 plt.yticks(fontsize = fontsize - 2)
50 ax.set_xlabel(
"X", fontsize = 17)
51 ax.set_ylabel(
"Y", fontsize = 17)
52 ax.set_title(prefixes[prefix], fontsize = fontsize)
55 plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
56 ax.tick_params(axis =
"y", which =
"minor")
57 ax.tick_params(axis =
"x", which =
"minor")
58 ax.set_axisbelow(
True)
61 plt.savefig(prefix +
".png")
63 sys.exit(
"Ambiguous file list exists.")
Visualization of the example output ⛓
- Test:
- test_pm_clusKmeans
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, September 1, 2012, 12:00 AM, National Institute for Fusion Studies, The University of Texas Austin
Definition at line 904 of file pm_clusKmeans.F90.