Return the predicted parallel Fork-Join speedup scaling behavior for simulations whose image contributions are stochastically accepted only from the first successful image starting from image ID 1.
More...
Return the predicted parallel Fork-Join speedup scaling behavior for simulations whose image contributions are stochastically accepted only from the first successful image starting from image ID 1.
This generic interface can be used to compute theoretical speedup gained by a Fork-Join parallel simulation where the probability of contribution of an image to the simulation is less than 1, meaning that image contributions are stochastic at each step of the Fork-Join cycle.
If the image contributions are inspected sequentially from the first image to last and only the first successful image contribution is kept in the simulation, then it can be shown that the contribution of the images to the parallel Fork-Join simulation follows a Cyclic Geometric Distribution.
See Amir Shahmoradi, Fatemeh Bagheri (2020). ParaDRAM: A Cross-Language Toolbox for Parallel High-Performance Delayed-Rejection Adaptive Metropolis Markov Chain Monte Carlo Simulations. for theoretical details.
- Parameters
-
| [in] | conProb | : The input scalar of,
-
type
real of kind any supported by the processor (e.g., RK, RK32, RK64, or RK128),
containing the average probability of contribution of each image to the Fork-Join simulation.
For example, conProb can be the effective acceptance rate in parallel Fork-Join Monte Carlo or MCMC sampling simulations where each image is tasked with returning a single proposal state for subsequent evaluation and if is first to be accepted accepted, for usage in the next simulation step.
|
| [in] | seqSecTime | : The input scalar of the same type and kind as conProb containing the time (in seconds) per image (or process or thread) of the inherently-sequential sections of the entire Fork-Join simulation.
|
| [in] | parSecTime | : The input scalar of the same type and kind as conProb containing the time (in seconds) per image (or process or thread) of the inherently-parallel sections of the entire Fork-Join simulation.
|
| [in] | comSecTime | : The input scalar of the same type and kind as conProb containing the time (in seconds) per image (or process or thread) of the pairwise inter-process communication sections of the entire Fork-Join simulation.
Assuming all rooter images communicate only with the main image and the entire communication takes overhead seconds, then comSecTime = overhead / (nproc - 1) with nproc representing the total number of images.
|
| [out] | scaling | : The output allocatable vector of the same type and kind as the input conProb containing the predicted speedup scaling of the stochastic Fork-Join simulation under a range of possible image (or thread or process) counts given in the output numproc.
On output, the vector scaling will be resized until the maximum speedup and the corresponding image count is identified in the scaling.
The ith element of scaling represents theoretically-predicted speedup if numproc(i) images (threads or processes) were used in the parallel Fork-Join simulation.
|
| [out] | numproc | : The output allocatable vector of type integer of default kind IK containing the number of images for the speedup reported in the corresponding element of scaling.
If the input argument comSecTime is positive, then numproc is simply a linear range of integers starting with 1 with a jump size of 1.
If the input argument comSecTime is positive, then numproc is simply a log(2)-linear range of integers starting with 1 with a jump size of 1.
|
| [in] | scalingMaxVal | : The output scalar of the same type and kind as the input conProb containing the maximum achievable speedup (corresponding to maxval(scaling)).
|
| [in] | scalingMaxLoc | : The output scalar of type integer of default kind IK containing the index of element of the output scaling containing scalingMaxVal (i.e., maxloc(scaling)).
|
| [out] | scalingMinLen | : The input positive scalar of type integer of default kind IK containing the minimum size of the output scaling.
(optional, default = int(2 / max(conProb, 0.001))) |
Possible calling interfaces ⛓
call setForkJoinScaling(conProb, seqSecTime, parSecTime, comSecTime, scaling, numproc, scalingMaxVal, scalingMaxLoc, scalingMinLen
= scalingMinLen)
!
Return the predicted parallel Fork-Join speedup scaling behavior for simulations whose image contribu...
This module contains procedures and generic interfaces for facilitating parallel computations or comp...
- Warning
- The condition
0 <= seqSecTime must hold for the corresponding input arguments.
The condition 0 <= parSecTime must hold for the corresponding input arguments.
The condition 0 <= comSecTime must hold for the corresponding input arguments.
The condition 0 <= scalingMinLen must hold for the corresponding input arguments.
The condition 0 <= conProb .and. conProb <= 1 must hold for the corresponding input arguments.
These conditions are verified only if the library is built with the preprocessor macro CHECK_ENABLED=1.
- See also
- pm_sampling
pm_distGeomCyclic
Amir Shahmoradi, Fatemeh Bagheri (2020). ParaDRAM: A Cross-Language Toolbox for Parallel High-Performance Delayed-Rejection Adaptive Metropolis Markov Chain Monte Carlo Simulations.
Example usage ⛓
11 type(display_type) :: disp
12 real(RKG) :: redshift
= 5.5_RKG
16 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
17 call disp%show(
"!Compute the Fisher z transformation of a random bounded variable.")
18 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
23 integer(IK),
allocatable :: numproc(:)
24 real,
allocatable :: scaling(:)
28 call disp%show(
"call setForkJoinScaling(conProb = .23, seqSecTime = 0., parSecTime = 1., comSecTime = .001, scaling = scaling, numproc = numproc, scalingMaxVal = maxval, scalingMaxLoc = maxloc)")
29 call setForkJoinScaling(conProb
= .
23, seqSecTime
= 0., parSecTime
= 1., comSecTime
= .
001, scaling
= scaling, numproc
= numproc, scalingMaxVal
= maxval, scalingMaxLoc
= maxloc)
47 real,
allocatable :: scaling(:)
48 integer(IK),
allocatable :: numproc(:)
49 call setForkJoinScaling(conProb
= .
23, seqSecTime
= 0., parSecTime
= 1., comSecTime
= .
001, scaling
= scaling, numproc
= numproc, scalingMaxVal
= maxval, scalingMaxLoc
= maxloc, scalingMinLen
= 32_IK)
50 if (
0 /= getErrTableWrite(
"setForkJoinScaling.csv",
reshape([
real(numproc), scaling], [
size(numproc),
2]), header
= "Processor Count,Scaling"))
error stop "Table writing failed."
Generate count evenly spaced points over the interval [x1, x2] if x1 < x2, or [x2,...
Generate and return a scalar or a contiguous array of rank 1 of length s1 of randomly uniformly distr...
Generate and return the iostat code resulting from writing the input table of rank 1 or 2 to the spec...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
This module contains procedures and generic interfaces for generating arrays with linear or logarithm...
This module contains classes and procedures for computing various statistical quantities related to t...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter RKD
The double precision real kind in Fortran mode. On most platforms, this is an 64-bit real kind.
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
integer, parameter RKS
The single-precision real kind in Fortran mode. On most platforms, this is an 32-bit real kind.
Generate and return an object of type display_type.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example output ⛓
7call setForkJoinScaling(conProb
= .
23, seqSecTime
= 0., parSecTime
= 1., comSecTime
= .
001, scaling
= scaling, numproc
= numproc, scalingMaxVal
= maxval, scalingMaxLoc
= maxloc)
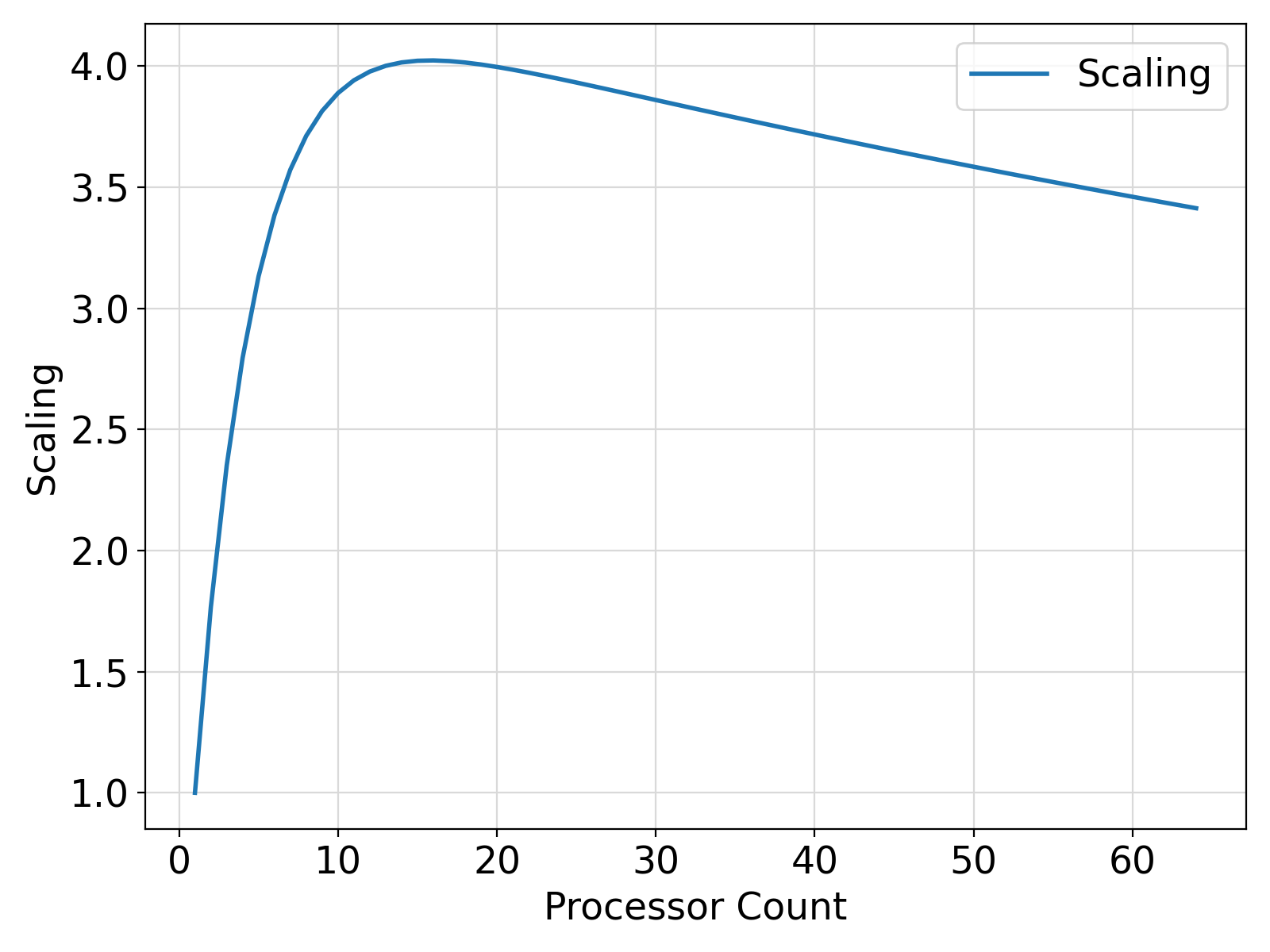
9+1.00000000,
+1.76687264,
+2.35178590,
+2.79578543,
+3.13124704,
+3.38341904,
+3.57183909,
+3.71154809,
+3.81408238,
+3.88827229,
+3.94086552,
+3.97701430,
+4.00064898,
+4.01476812,
+4.02165937,
+4.02306509,
+4.02031755,
+4.01442909,
+4.00617361,
+3.99614286,
+3.98478413,
+3.97244120,
+3.95937586,
+3.94578743,
+3.93182707,
+3.91761041,
+3.90322590,
+3.88874030,
+3.87420368,
+3.85965514,
+3.84512305,
+3.83063030,
+3.81619239,
+3.80182147,
+3.78752732,
+3.77331614,
+3.75919294,
+3.74516082,
+3.73122287,
+3.71738029,
+3.70363355,
+3.68998289,
+3.67642927,
+3.66297197,
+3.64961052,
+3.63634515,
+3.62317395,
+3.61009741,
+3.59711385,
+3.58422279,
+3.57142329,
+3.55871487,
+3.54609632,
+3.53356671,
+3.52112484,
+3.50877047,
+3.49650216,
+3.48431945,
+3.47222137,
+3.46020651,
+3.44827533,
+3.43642521,
+3.42465687,
+3.41296887,
+3.40135980,
+3.38982987,
+3.37837839,
+3.36700320,
+3.35570431,
+3.34448123,
+3.33333325,
+3.32225871
11+1,
+2,
+3,
+4,
+5,
+6,
+7,
+8,
+9,
+10,
+11,
+12,
+13,
+14,
+15,
+16,
+17,
+18,
+19,
+20,
+21,
+22,
+23,
+24,
+25,
+26,
+27,
+28,
+29,
+30,
+31,
+32,
+33,
+34,
+35,
+36,
+37,
+38,
+39,
+40,
+41,
+42,
+43,
+44,
+45,
+46,
+47,
+48,
+49,
+50,
+51,
+52,
+53,
+54,
+55,
+56,
+57,
+58,
+59,
+60,
+61,
+62,
+63,
+64,
+65,
+66,
+67,
+68,
+69,
+70,
+71,
+72
Postprocessing of the example output ⛓
5examname = os.path.basename(os.getcwd())
9import matplotlib.pyplot
as plt
15df = pd.read_csv(examname +
".csv", delimiter =
",")
17fig = plt.figure(figsize = 1.25 * np.array([6.4, 4.8]), dpi = 200)
20for colname
in df.columns[1:]:
21 plt.plot( df.values[:, 0]
28for colname
in list(df.columns[1:]): labels.append(colname)
35ax.set_xlabel(df.columns[0], fontsize = fontsize)
36ax.set_ylabel(df.columns[1], fontsize = fontsize)
38plt.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
39ax.tick_params(axis =
"y", which =
"minor")
40ax.tick_params(axis =
"x", which =
"minor")
41plt.xticks(fontsize = fontsize)
42plt.yticks(fontsize = fontsize)
45plt.savefig(examname +
".png")
Visualization of the example output ⛓
- Test:
- test_pm_parallelism
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, Tuesday March 7, 2017, 4:13 AM, Institute for Computational Engineering and Sciences (ICES), The University of Texas Austin
Definition at line 229 of file pm_parallelism.F90.