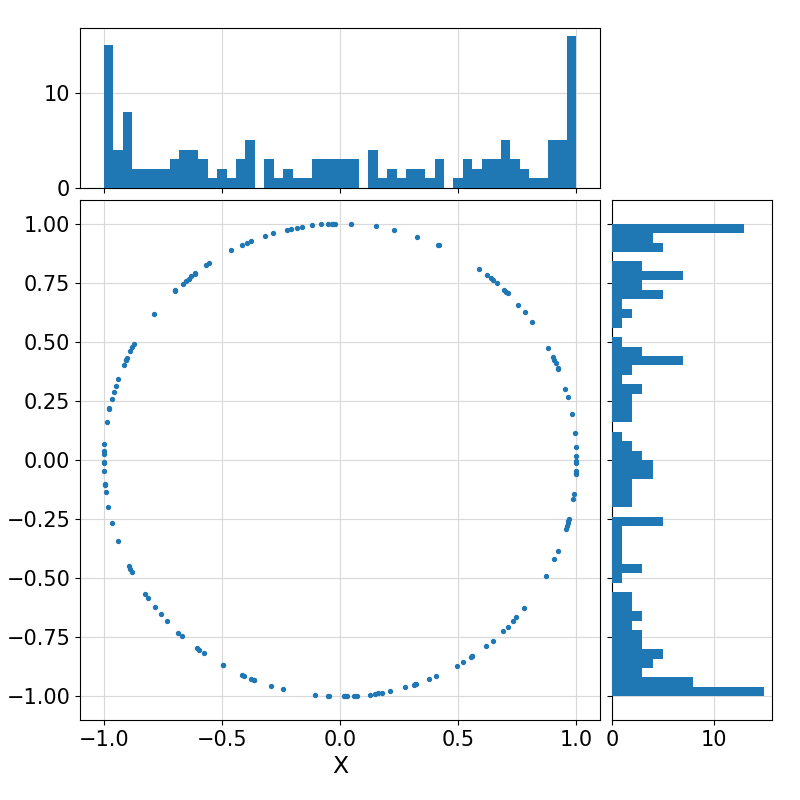
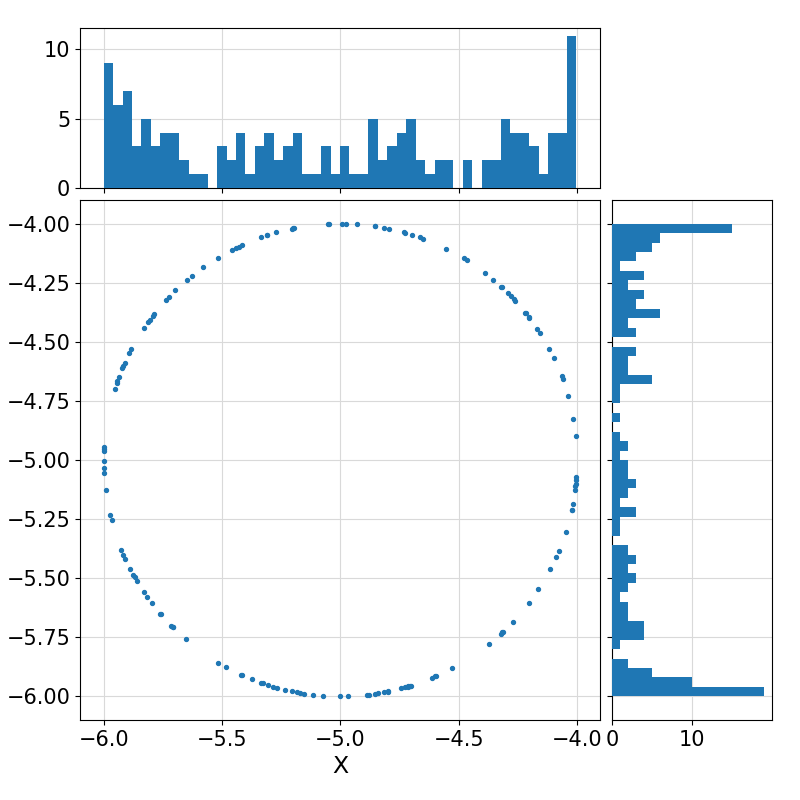
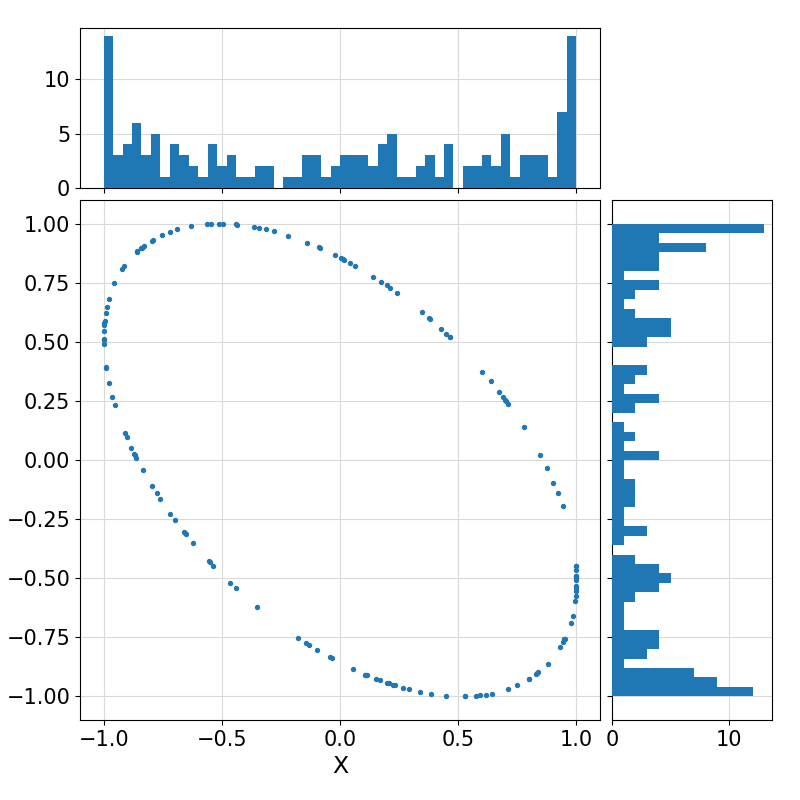
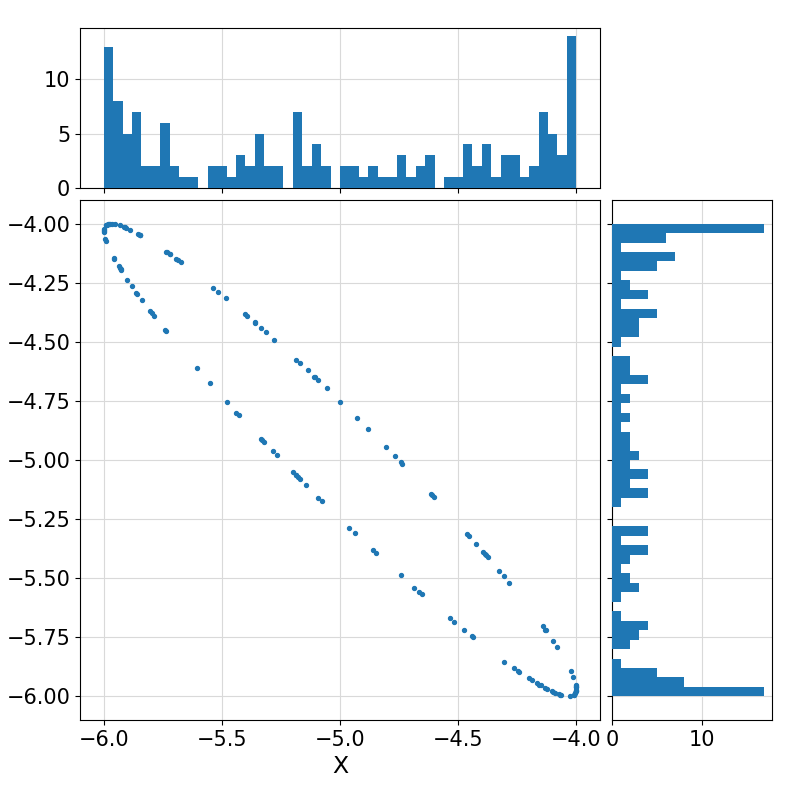
Return a (collection) of random vector(s) of size ndim uniformly distributed on the surface of an n-sphere, optionally with the specified input mean(1:ndim) and optionally affine-transformed to a non-uniform distribution on the surface of an (n+1)-ellipsoid represented by the Cholesky Factorization of its Gramian matrix.
- Parameters
-
| [in,out] | rng | : The input/output scalar that can be an object of,
-
type rngf_type, implying the use of intrinsic Fortran uniform RNG.
-
type xoshiro256ssw_type, implying the use of xoshiro256** uniform RNG.
(optional, default = rngf_type.) |
| [out] | rand | : The output contiguous vector of shape (1:ndim) or matrix of shape (1:ndim, 1:nsam) of
-
type
real of kind any supported by the processor (e.g., RK, RK32, RK64, or RK128),
containing the random output vector.
|
| [in] | mean | : The input contiguous vector of shape (1:ndim), of the same type and kind as the output rand, representing the center of the sphere.
(optional, default = [(0., i = 1, size(rand))]. It must be present if the input argument chol is missing.) |
| [in] | chol | : The input contiguous matrix of shape (ndim, ndim) whose specified triangular subset contains the Cholesky Factorization of the Gramian matrix of the corresponding hyper-ellipsoid on which the output random vectors must be distributed proportional to the ellipsoidal surface curvature.
(optional, the default is the Identity matrix of rank ndim. It must be present if and only if the input argument subset is also present.) |
| [in] | subset | : The input scalar constant that can be any of the following:
-
the constant uppDia or an object of type uppDia_type implying that the upper-diagonal triangular block of the input
chol must be used while the lower subset is not referenced.
-
the constant lowDia or an object of type lowDia_type implying that the lower-diagonal triangular block of the input
chol must be used while the upper subset is not referenced.
This argument is merely a convenience to differentiate the different procedure functionalities within this generic interface.
(optional. It must be present if and only if the input argument chol is present.) |
Possible calling interfaces ⛓
call setUnifSphereRand(rng, rand(
1:ndim), mean(
1:ndim), chol(
1:ndim,
1:ndim), subset)
call setUnifSphereRand(rand(
1:ndim,
1:nsam), mean(
1:ndim), chol(
1:ndim,
1:ndim), subset)
call setUnifSphereRand(rng, rand(
1:ndim,
1:nsam), mean(
1:ndim), chol(
1:ndim,
1:ndim), subset)
Return a (collection) of random vector(s) of size ndim uniformly distributed on the surface of an -sp...
This module contains classes and procedures for computing various statistical quantities related to t...
- Warning
- The condition
size(mean, 1) == size(rand, 1) must hold for the corresponding input arguments.
The condition all(shape(chol) == size(rand, 1)) must hold for the corresponding input arguments.
These conditions are verified only if the library is built with the preprocessor macro CHECK_ENABLED=1.
- See also
- getNormRand
setNormRand
getUnifRand
setUnifRand
getUnifSphereRand
setUnifSphereRand
getUnifEllRand
setUnifEllRand
getUnifParRand
setUnifParRand
Example usage ⛓
12 integer(IK),
parameter :: NDIM
= 2_IK
13 real(RK) :: mean(NDIM), gramian(NDIM, NDIM), chol(NDIM, NDIM), rand(NDIM)
16 type(display_type) :: disp
20 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
21 call disp%show(
"! Multivariate Uniform Ellipsoidal random vector with given mean and Gramian matrix specified via the Cholesky Lower Triangle.")
22 call disp%show(
"!%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%")
26 call disp%show(
"call setUnifSphereRand(rand)")
35 call disp%show(
"call setUnifSphereRand(rand, mean)")
42 call disp%show(
"gramian = getCovRand(mold = 1., ndim = ndim)")
44 call disp%show(
"chol = getMatChol(gramian, lowDia)")
48 call disp%show(
"call setUnifSphereRand(rand, chol, lowDia)")
59 call disp%show(
"call setUnifSphereRand(rand, mean, chol, lowDia)")
70 integer(IK) :: fileUnit, i
71 integer(IK),
parameter :: NVEC
= 150_IK
72 open(newunit
= fileUnit, file
= "setUnifSphereRand.RK.txt")
75 write(fileUnit,
"(*(g0,:,','))") rand
78 open(newunit
= fileUnit, file
= "setUnifSphereRandMean.RK.txt")
81 write(fileUnit,
"(*(g0,:,','))") rand
84 open(newunit
= fileUnit, file
= "setUnifSphereRandChol.RK.txt")
88 write(fileUnit,
"(*(g0,:,','))") rand
91 open(newunit
= fileUnit, file
= "setUnifSphereRandMeanChol.RK.txt")
95 write(fileUnit,
"(*(g0,:,','))") rand
Generate and return a random positive-definite (correlation or covariance) matrix using the Gram meth...
This is a generic method of the derived type display_type with pass attribute.
This is a generic method of the derived type display_type with pass attribute.
Generate and return the upper or the lower Cholesky factorization of the input symmetric positive-def...
This module contains classes and procedures for generating random matrices distributed on the space o...
This module contains classes and procedures for input/output (IO) or generic display operations on st...
type(display_type) disp
This is a scalar module variable an object of type display_type for general display.
This module defines the relevant Fortran kind type-parameters frequently used in the ParaMonte librar...
integer, parameter RK
The default real kind in the ParaMonte library: real64 in Fortran, c_double in C-Fortran Interoperati...
integer, parameter LK
The default logical kind in the ParaMonte library: kind(.true.) in Fortran, kind(....
integer, parameter IK
The default integer kind in the ParaMonte library: int32 in Fortran, c_int32_t in C-Fortran Interoper...
integer, parameter SK
The default character kind in the ParaMonte library: kind("a") in Fortran, c_char in C-Fortran Intero...
This module contains procedures and generic interfaces for computing the Cholesky factorization of po...
Generate and return an object of type display_type.
Example Unix compile command via Intel ifort compiler ⛓
3ifort -fpp -standard-semantics -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example Windows Batch compile command via Intel ifort compiler ⛓
2set PATH=..\..\..\lib;%PATH%
3ifort /fpp /standard-semantics /O3 /I:..\..\..\include main.F90 ..\..\..\lib\libparamonte*.lib /exe:main.exe
Example Unix / MinGW compile command via GNU gfortran compiler ⛓
3gfortran -cpp -ffree-line-length-none -O3 -Wl,-rpath,../../../lib -I../../../inc main.F90 ../../../lib/libparamonte* -o main.exe
Example output ⛓
9-0.40268694527372084,
-0.91533776503874209
15-4.4621357081631841,
-5.8430314368793610
21+1.0000000000000000,
+0.0000000000000000
22+0.52618491649627686,
+0.85037017448391627
25+0.72814742900816354,
+0.96600138141037184
29-5.0000000000000000,
-5.0000000000000000
31+1.0000000000000000,
+0.0000000000000000
32+0.52618491649627686,
+0.85037017448391627
35-5.0960189434522167,
-4.2040826748924758
Postprocessing of the example output ⛓
3import matplotlib.pyplot
as plt
14 pattern =
"*." + kind +
".txt"
15 fileList = glob.glob(pattern)
19 df = pd.read_csv(file, delimiter =
",", header =
None)
22 left, width = 0.1, 0.65
23 bottom, height = 0.1, 0.65
27 fig = plt.figure(figsize = (8, 8))
29 plt.rcParams.update({
'font.size': fontsize - 2})
30 ax = fig.add_axes([left, bottom, width, height])
31 ax_histx = fig.add_axes([left, bottom + height + spacing, width, 0.2], sharex = ax)
32 ax_histy = fig.add_axes([left + width + spacing, bottom, 0.2, height], sharey = ax)
34 for axes
in [ax, ax_histx, ax_histy]:
35 axes.grid(visible =
True, which =
"both", axis =
"both", color =
"0.85", linestyle =
"-")
36 axes.tick_params(axis =
"y", which =
"minor")
37 axes.tick_params(axis =
"x", which =
"minor")
40 ax_histy.tick_params(axis =
"y", labelleft =
False)
41 ax_histx.tick_params(axis =
"x", labelbottom =
False)
44 ax.scatter ( df.values[:, 0]
50 ax_histx.hist(df.values[:, 0], bins = 50, zorder = 1000)
51 ax_histy.hist(df.values[:, 1], bins = 50, orientation =
"horizontal", zorder = 1000)
53 ax.set_xlabel(
"X", fontsize = 17)
54 ax.set_ylabel(
"Y", fontsize = 17)
57 plt.savefig(file.replace(
".txt",
".png"))
Visualization of the example output ⛓
- Test:
- test_pm_distUnifSphere
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
-
If you use any parts or concepts from this library to any extent, please acknowledge the usage by citing the relevant publications of the ParaMonte library.
-
If you regenerate any parts/ideas from this library in a programming environment other than those currently supported by this ParaMonte library (i.e., other than C, C++, Fortran, MATLAB, Python, R), please also ask the end users to cite this original ParaMonte library.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
- Copyright
- Computational Data Science Lab
- Author:
- Amir Shahmoradi, April 23, 2017, 12:36 AM, Institute for Computational Engineering and Sciences (ICES), University of Texas at Austin
Definition at line 2576 of file pm_distUnifSphere.F90.