|
ParaMonte MATLAB 3.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
|
ParaMonte MATLAB 3.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
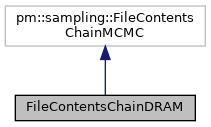
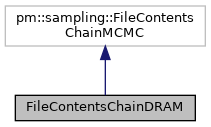
This is the base class for generating objects that contain the contents of a chain file generated by a sampler of superclass pm.sampling.Paradram.
More...
Public Member Functions | |
| function | FileContentsChainDRAM (in file, in silent, in sep, in format) |
| Return a scalar object of class pm.sampling.FileContentsChainDRAM. More... | |
This is the base class for generating objects that contain the contents of a chain file generated by a sampler of superclass pm.sampling.Paradram.
This class is meant to be primarily internally used by the ParaMonte MATLAB library samplers.
This class merely adds a number of visualizations to its superclass which are specific to the pm.sampling.Paradram sampler.
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
Definition at line 28 of file FileContentsChainDRAM.m.
| function FileContentsChainDRAM::FileContentsChainDRAM | ( | in | file, |
| in | silent, | ||
| in | sep, | ||
| in | format | ||
| ) |
Return a scalar object of class pm.sampling.FileContentsChainDRAM.
This is the constructor of the class pm.sampling.FileContentsChainDRAM.
| [in] | file | : The input scalar MATLAB string containing the path or web address to an external file. |
| [in] | silent | : See the corresponding argument of the superclass constructor pm.sampling.FileContentsChainMCMC::FileContentsChainMCMC. (optional. The default is set by the superclass constructor pm.sampling.FileContentsChainMCMC::FileContentsChainMCMC.) |
| [in] | sep | : See the corresponding argument of the superclass constructor pm.sampling.FileContentsChainMCMC::FileContentsChainMCMC. (optional. The default is set by the superclass constructor pm.sampling.FileContentsChainMCMC::FileContentsChainMCMC.) |
| [in] | format | : The input scalar MATLAB string containing the reading format of the Markov chain:
"compact".) |
self : The output scalar object of class pm.sampling.FileContentsChainDRAM.
Possible calling interfaces ⛓
Example usage ⛓
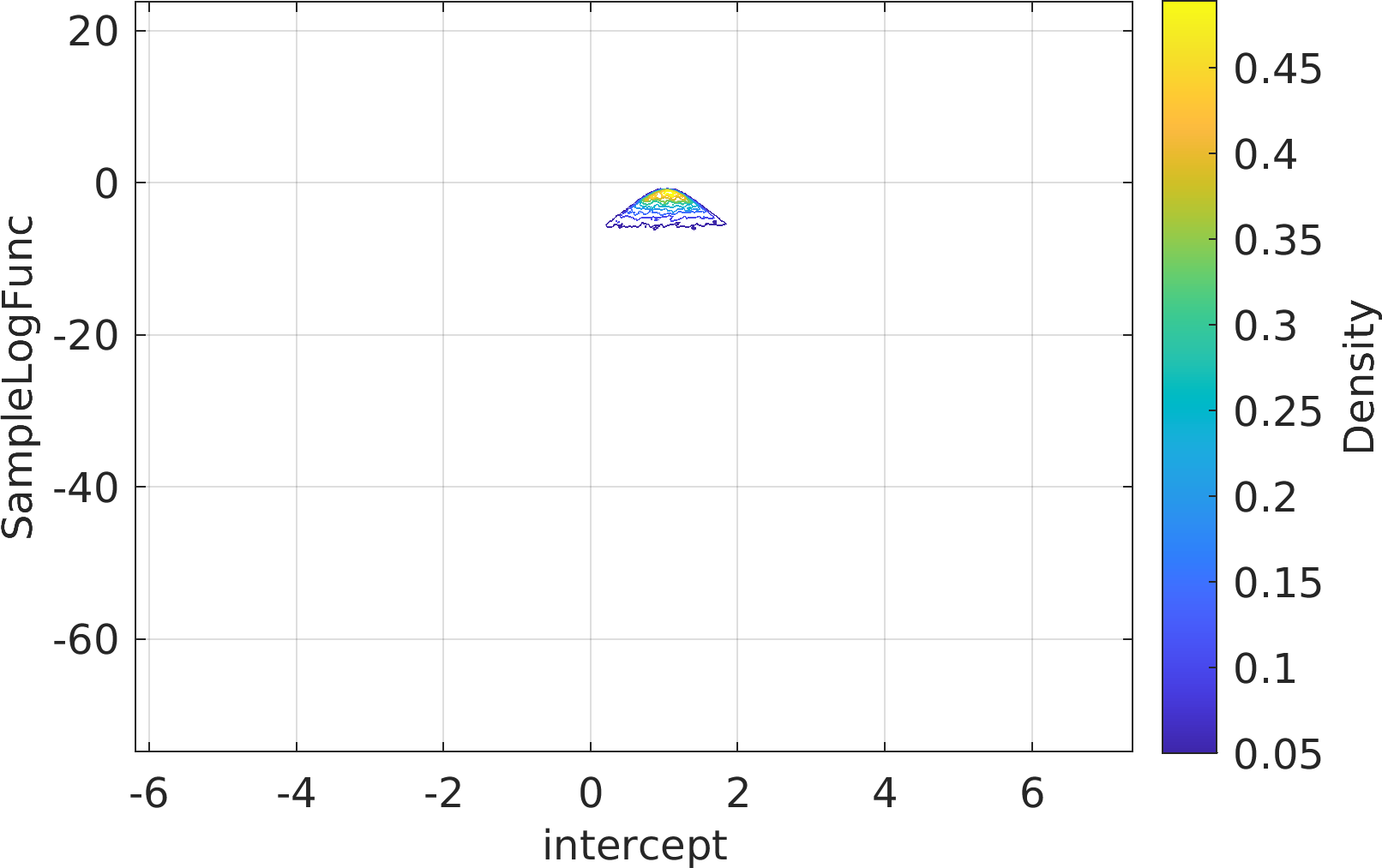
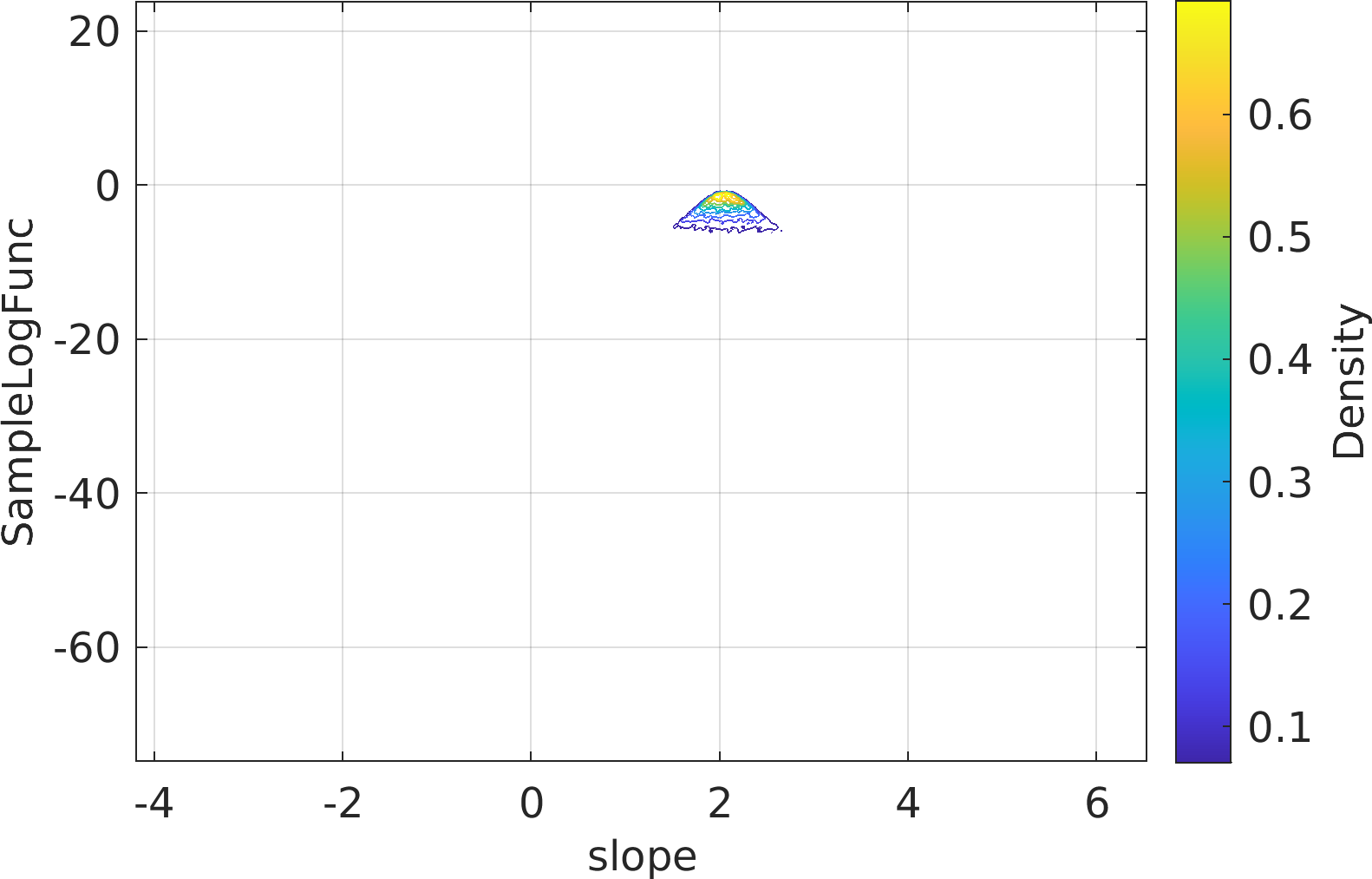
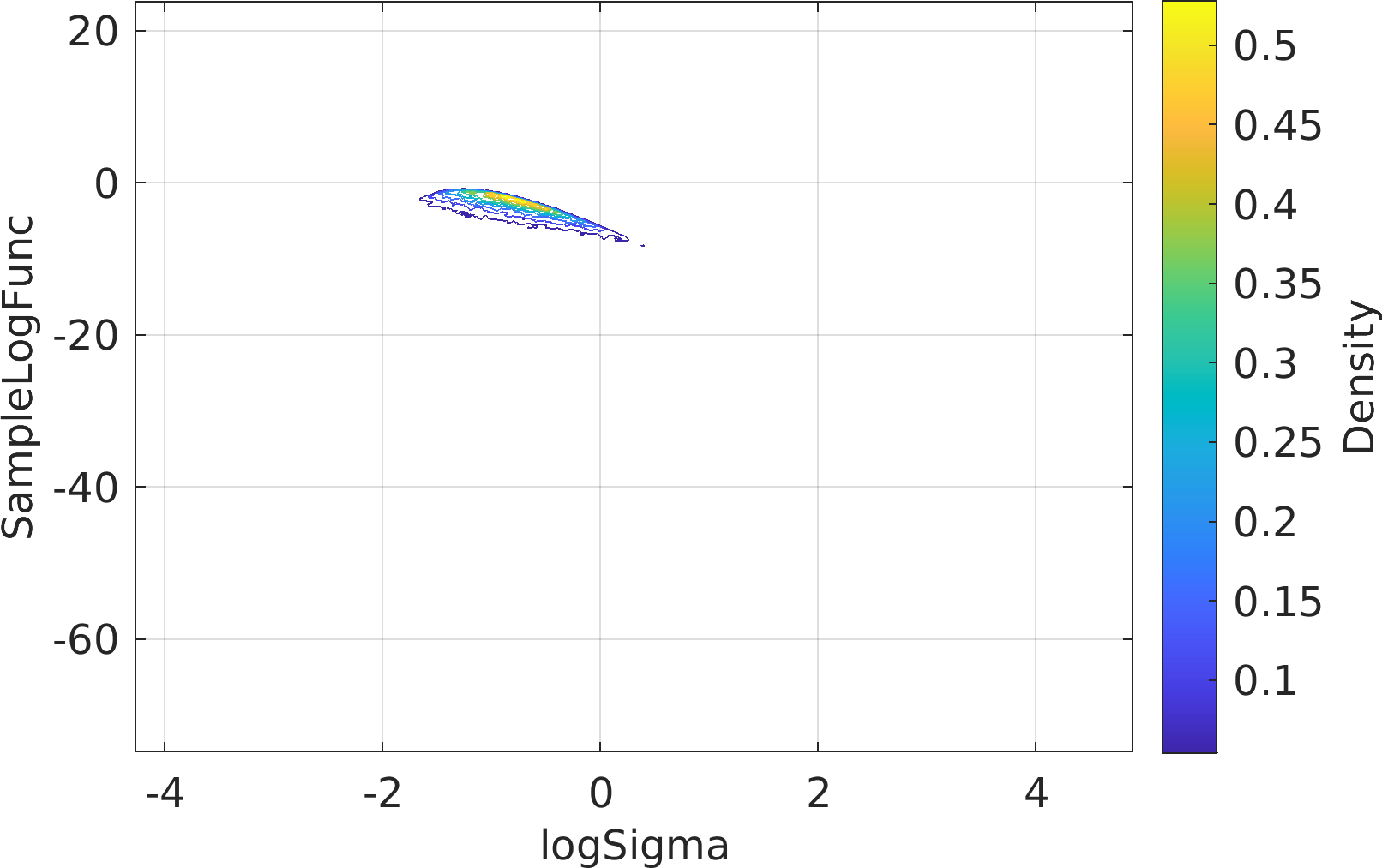
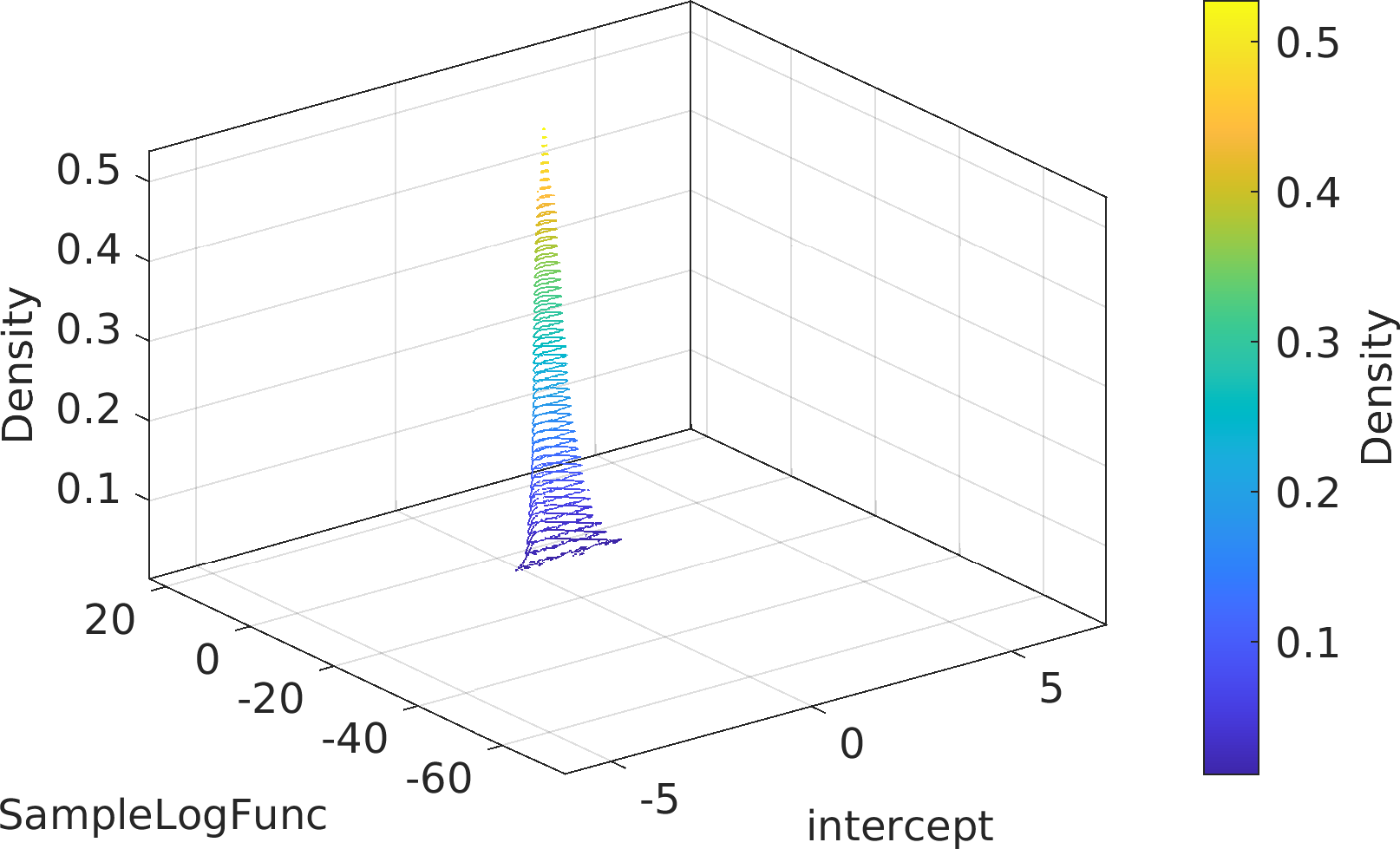
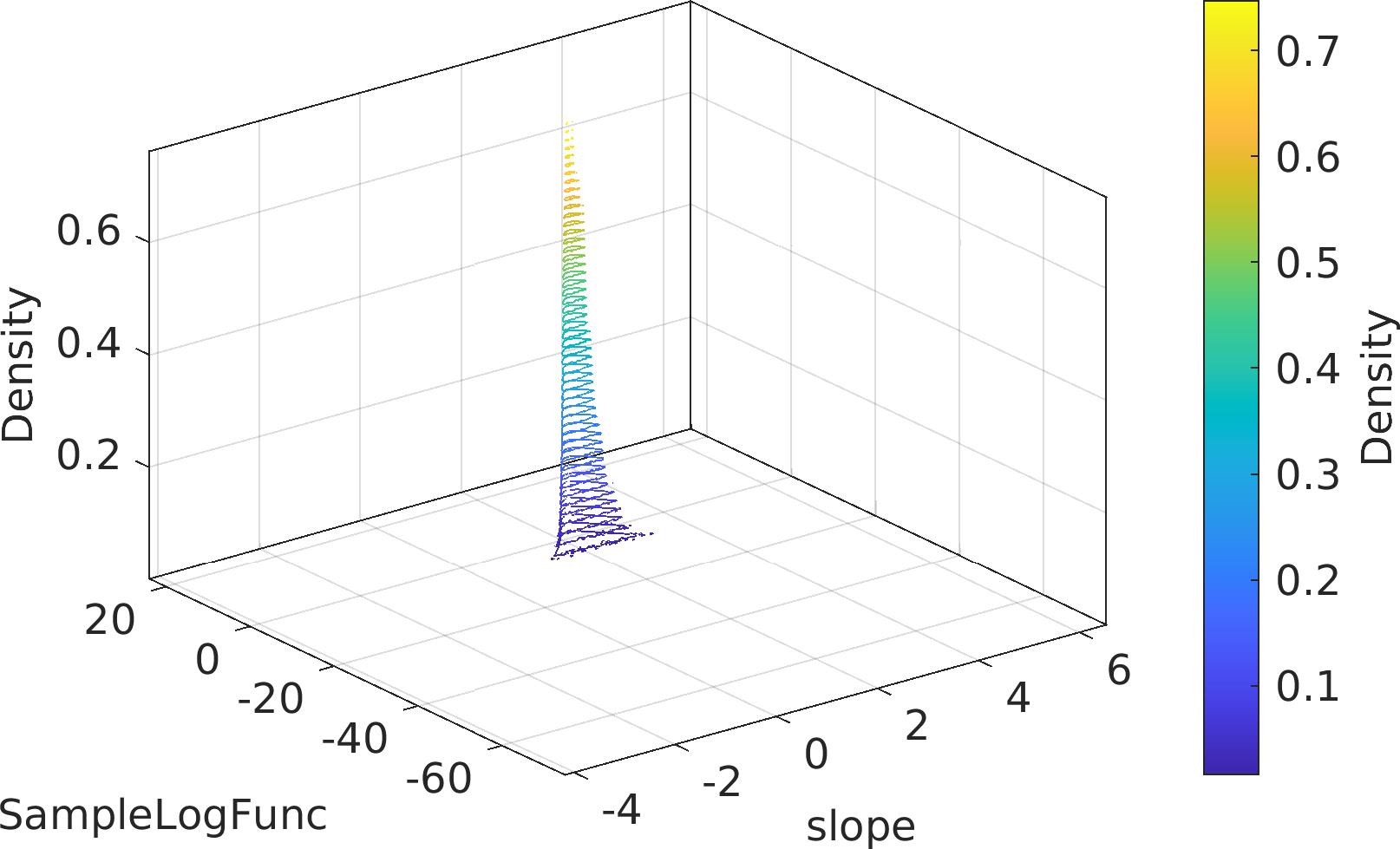
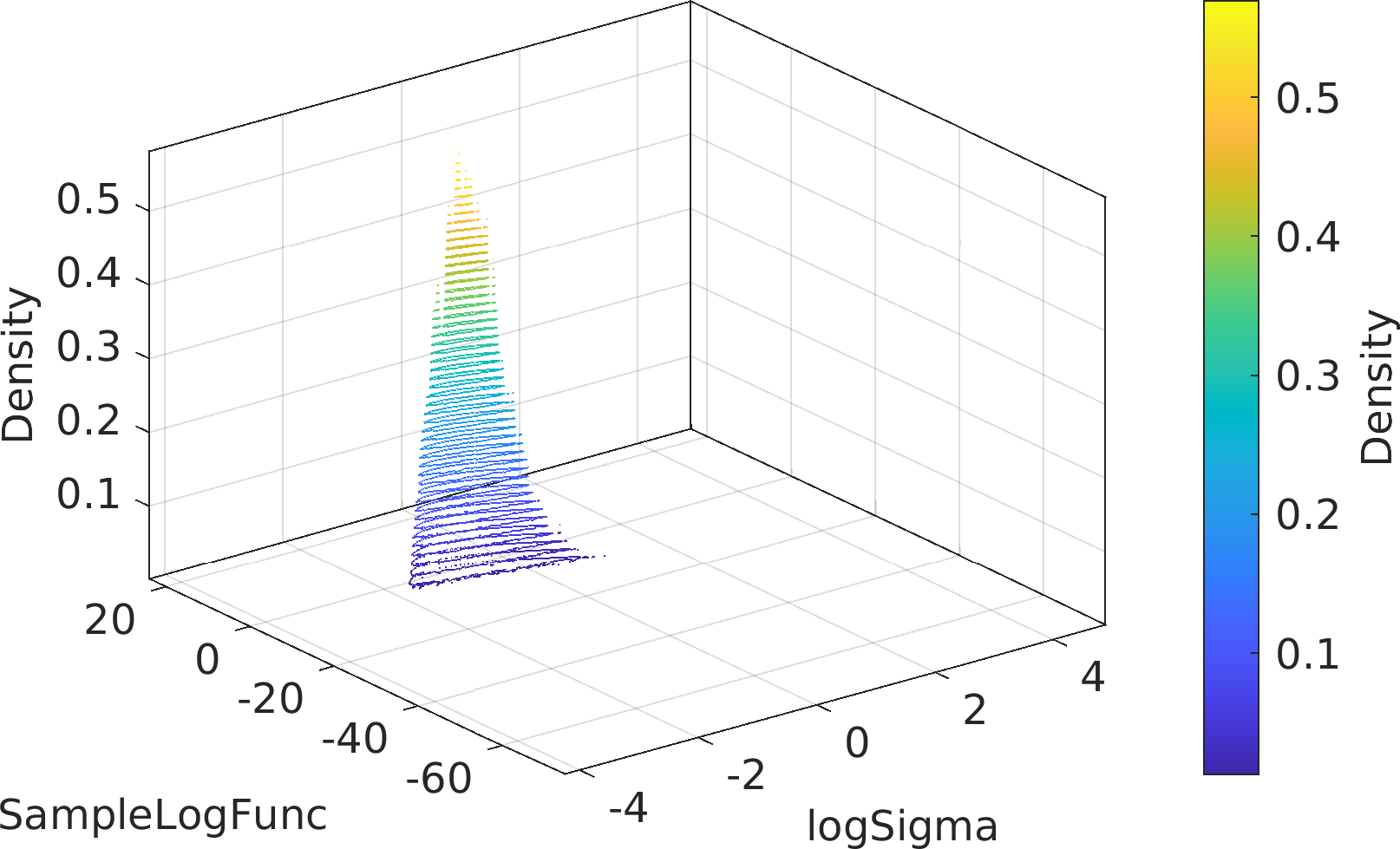
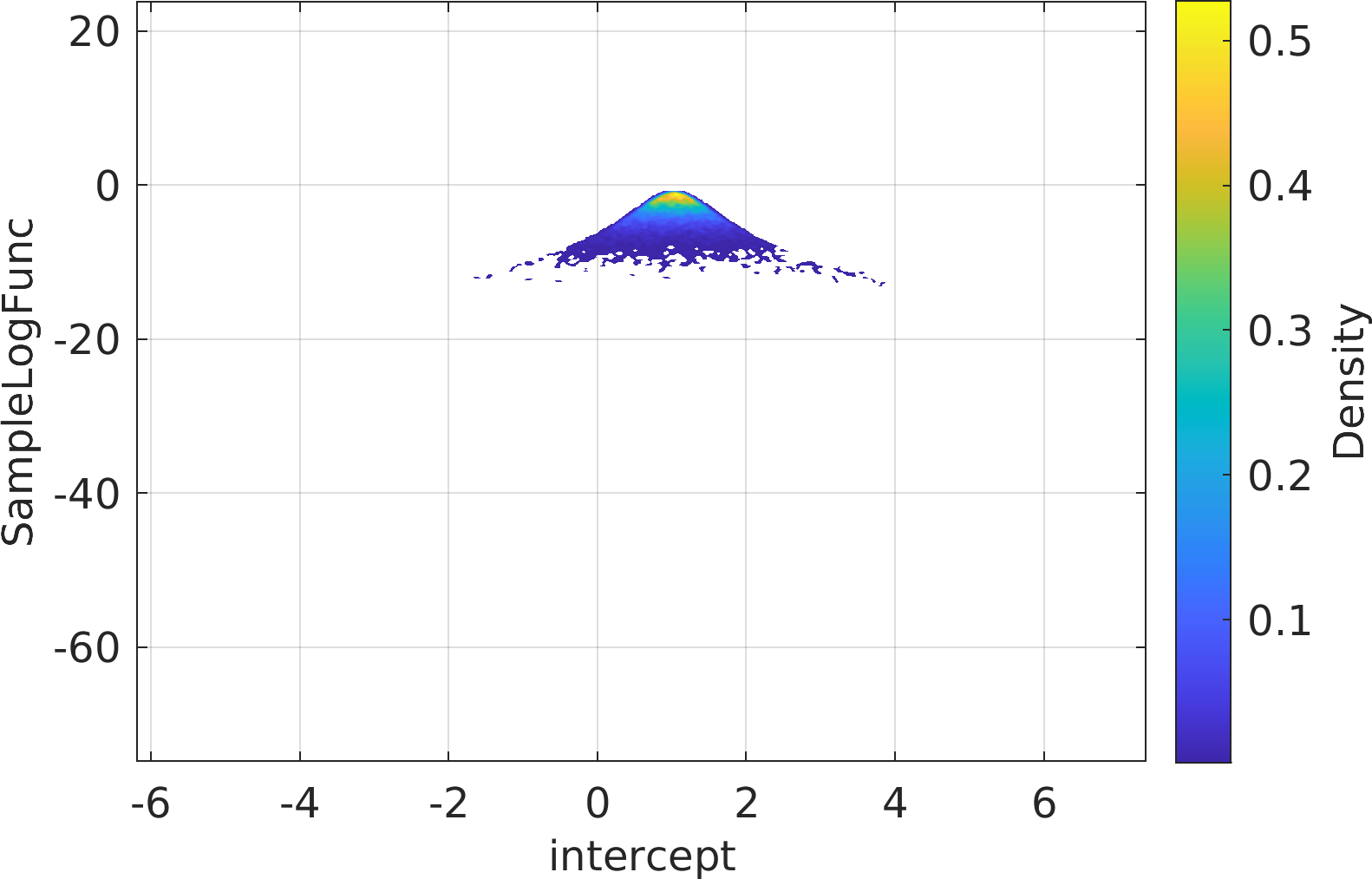
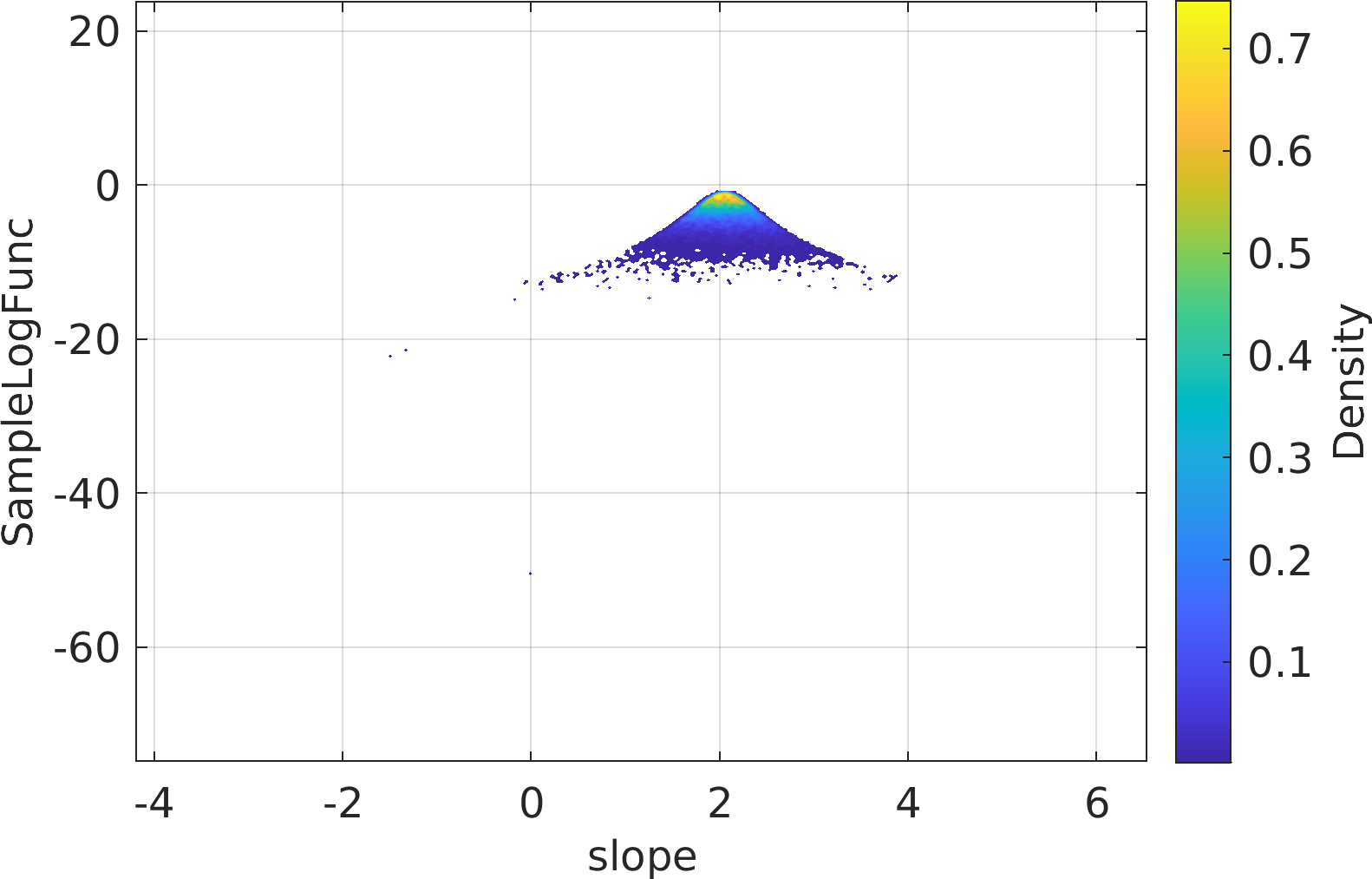
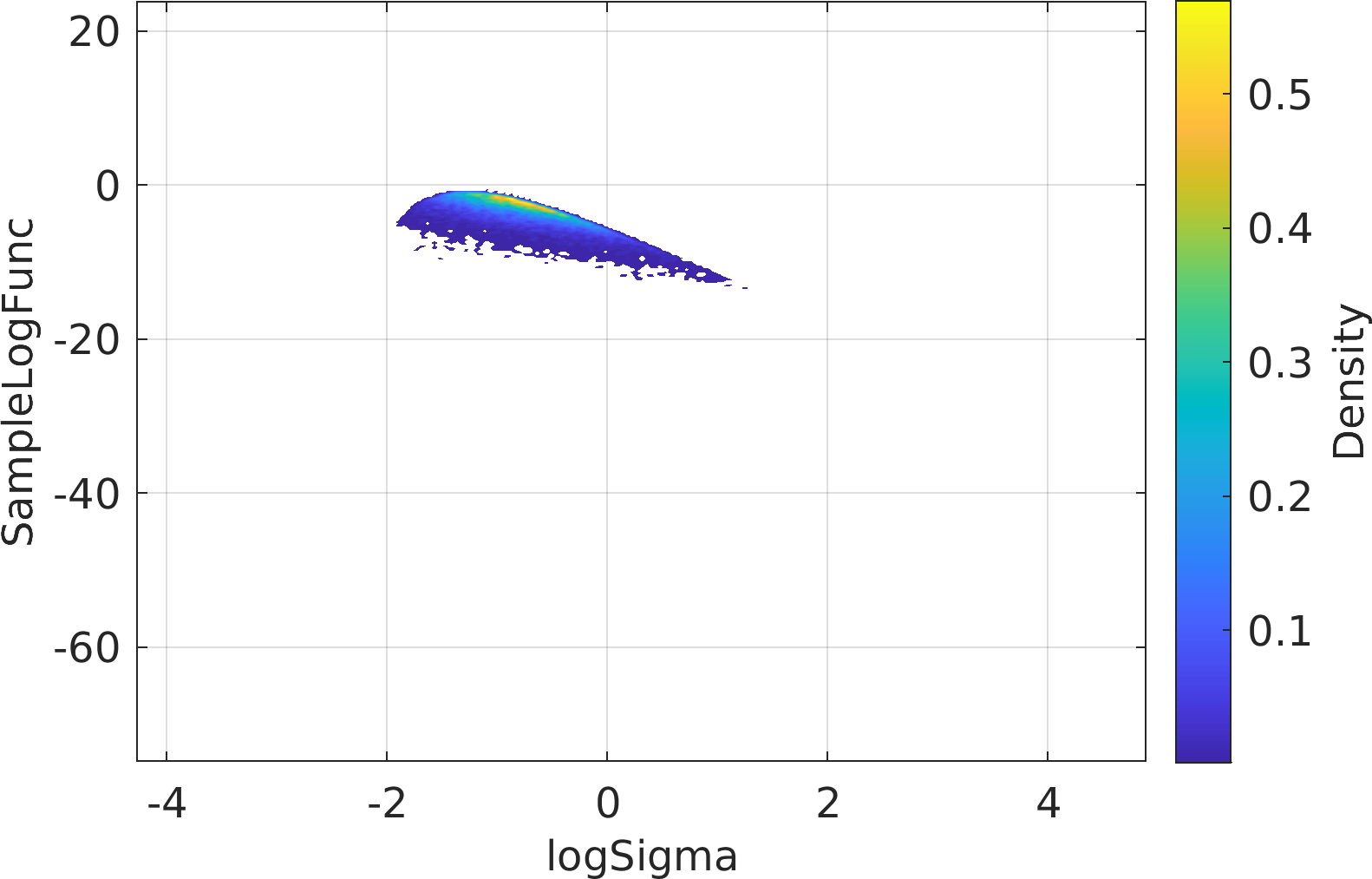
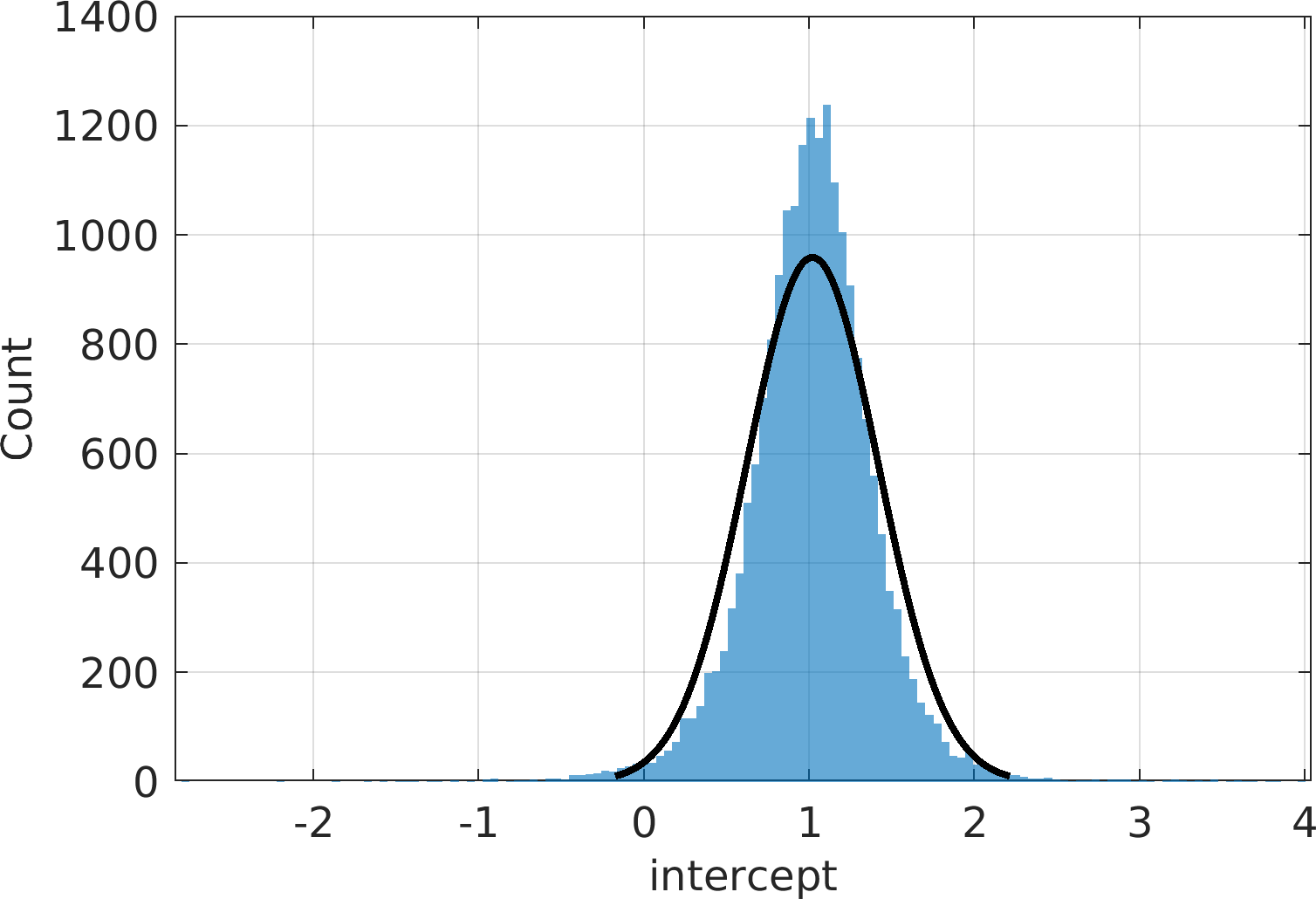
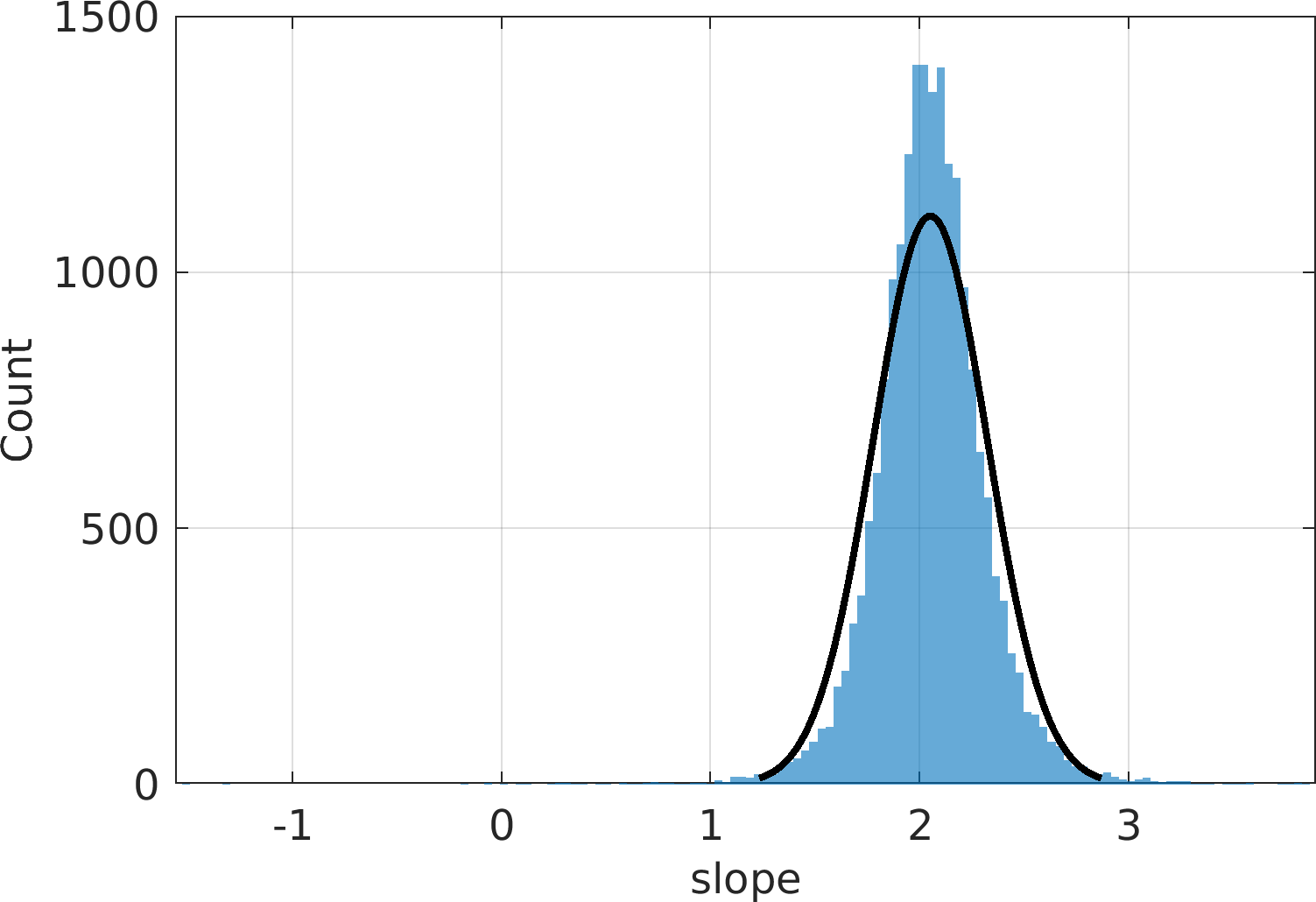
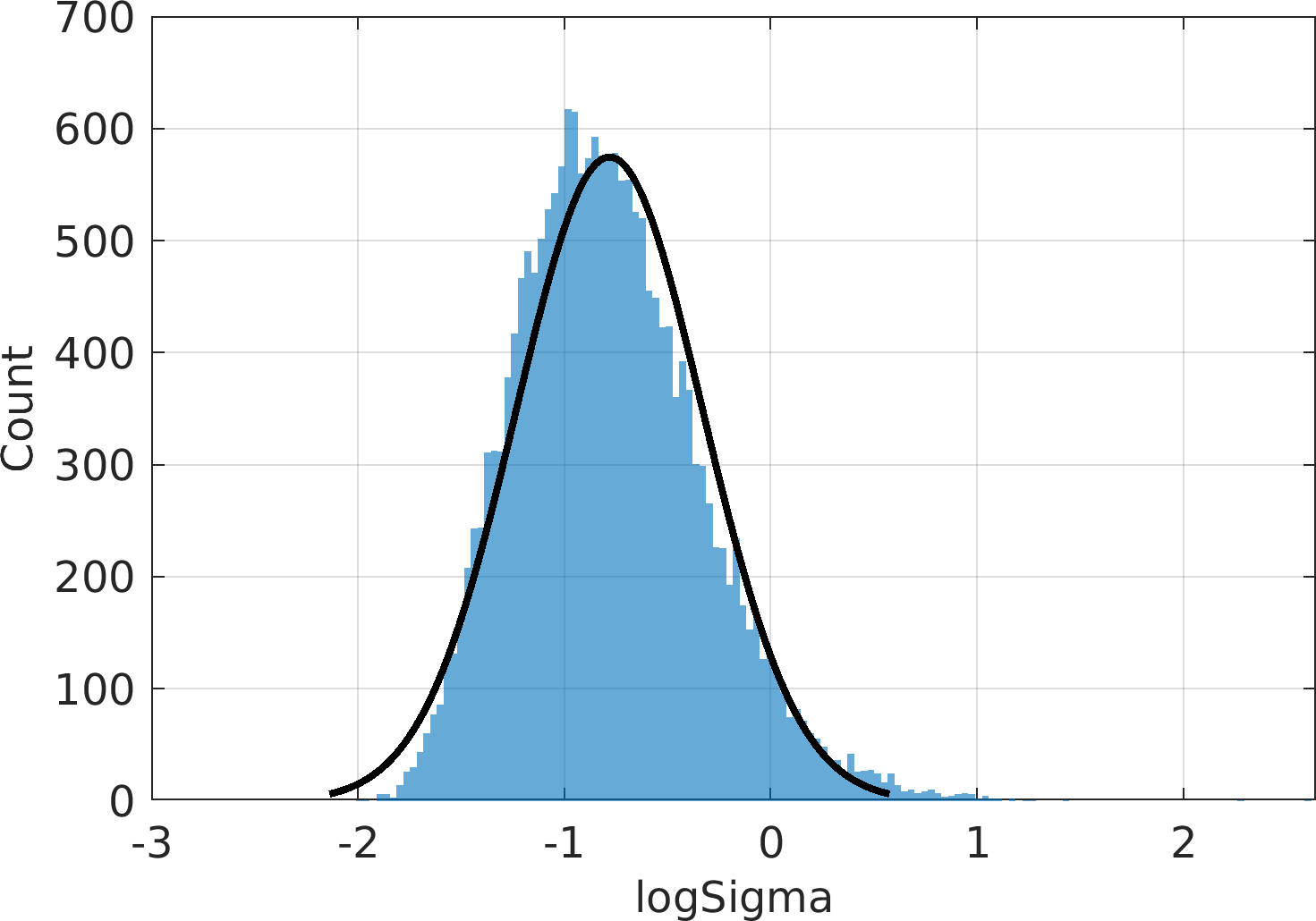
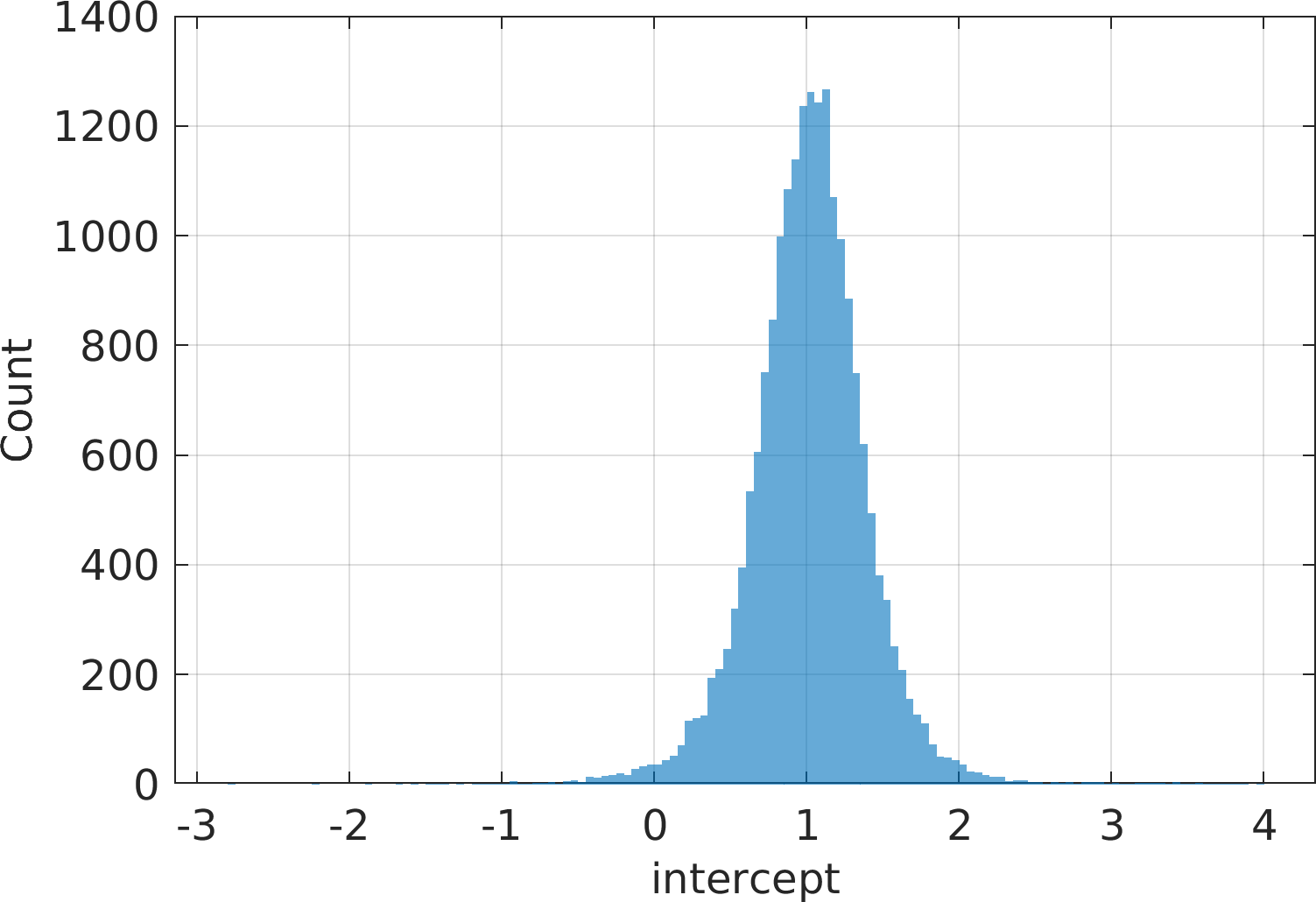
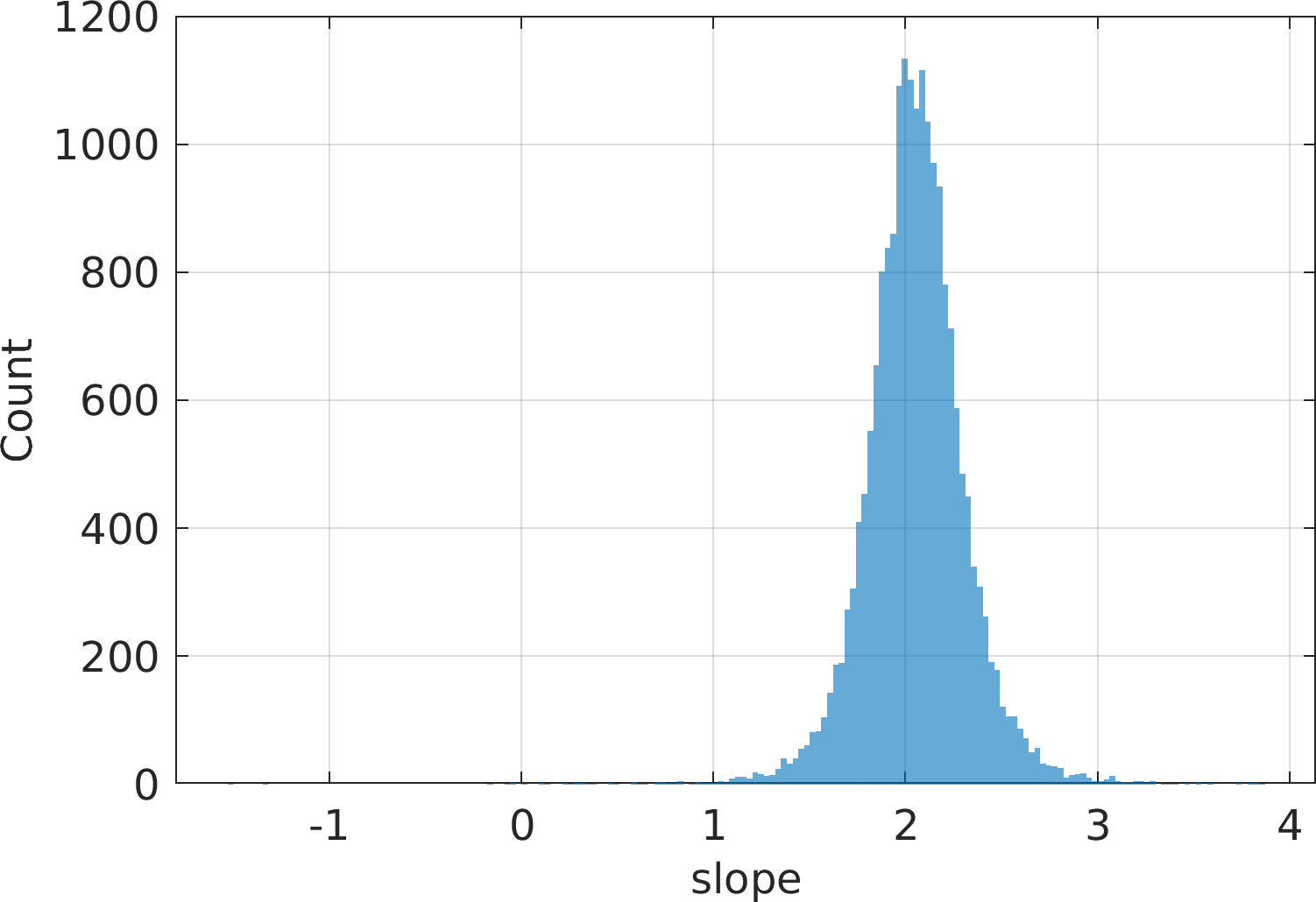
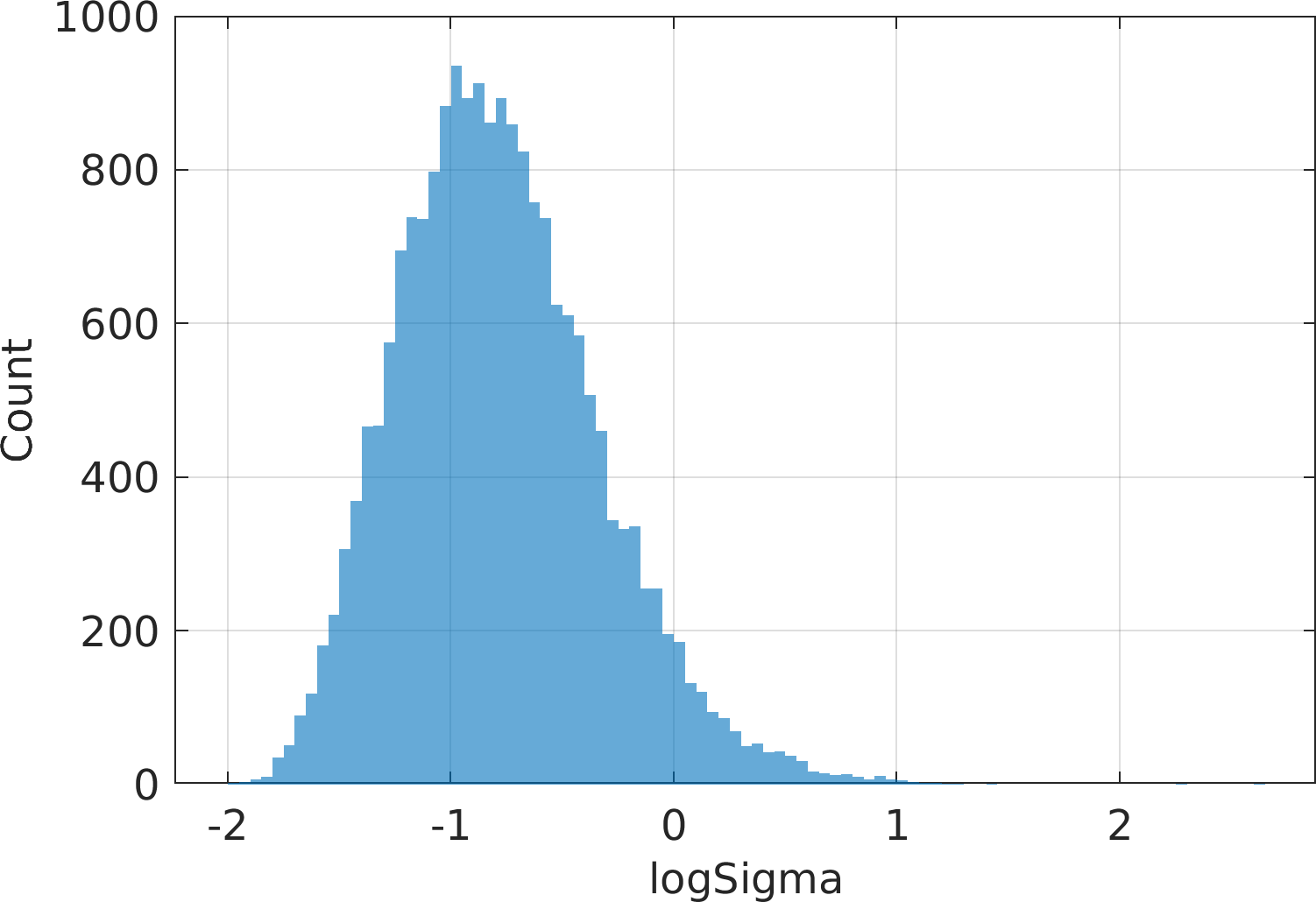
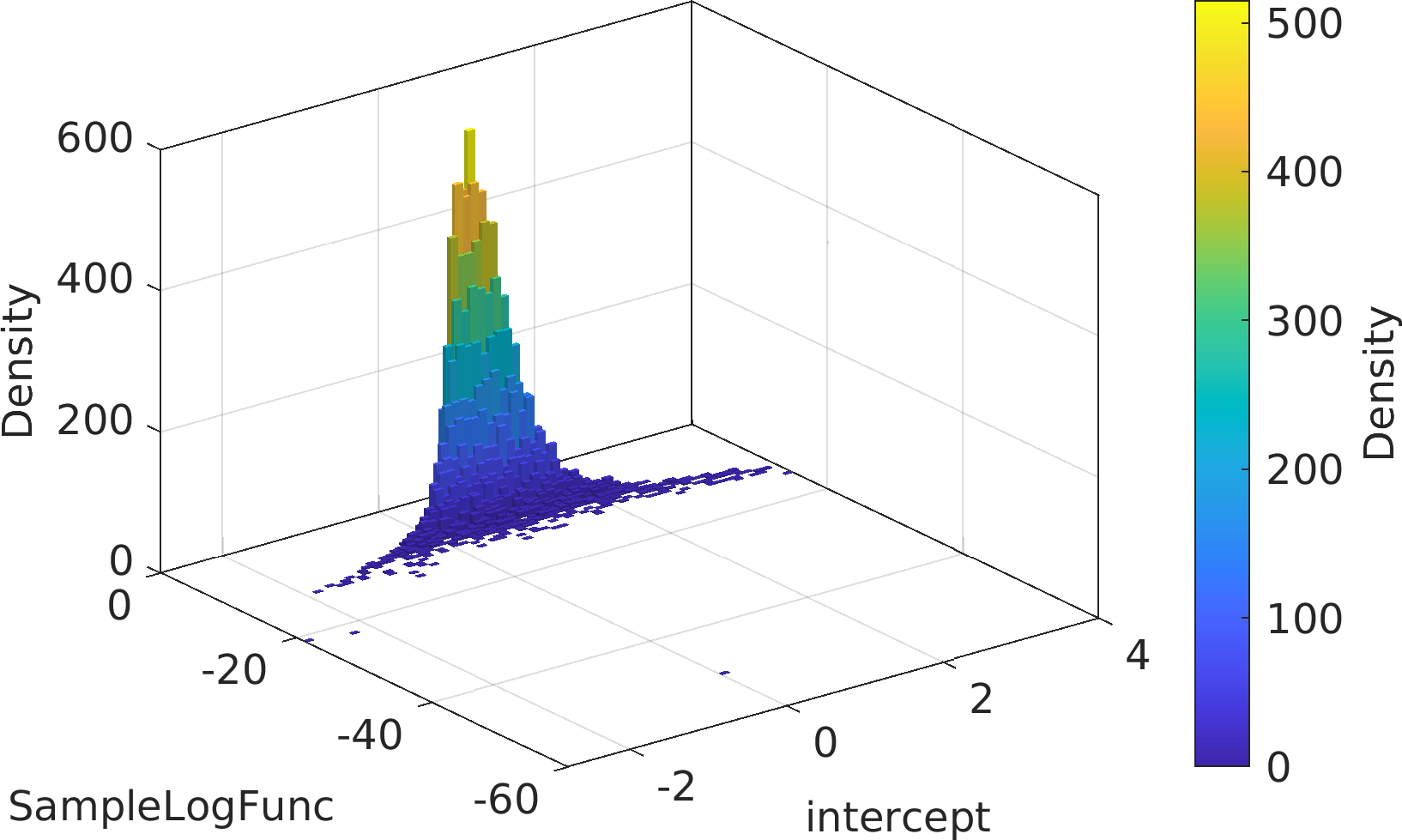
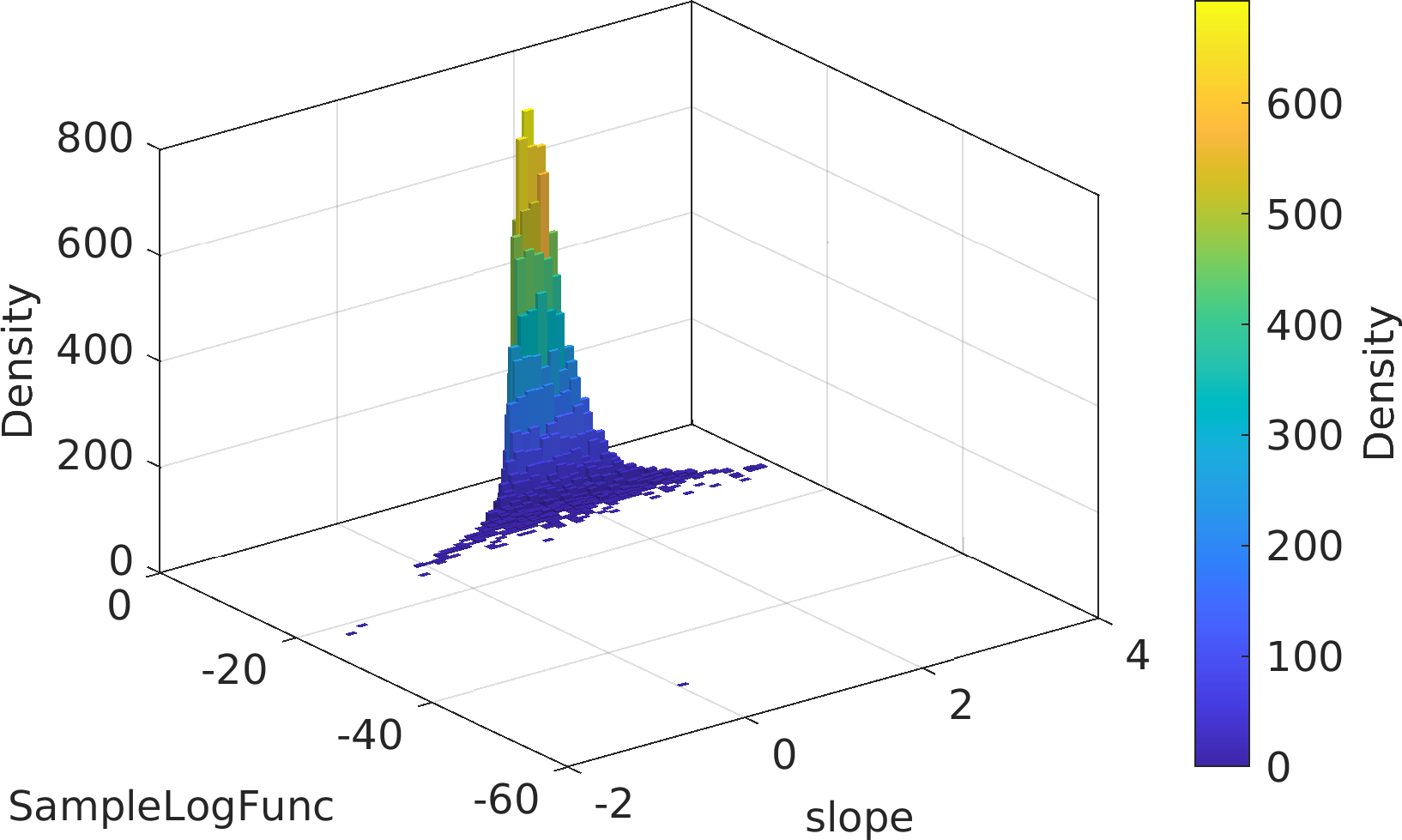
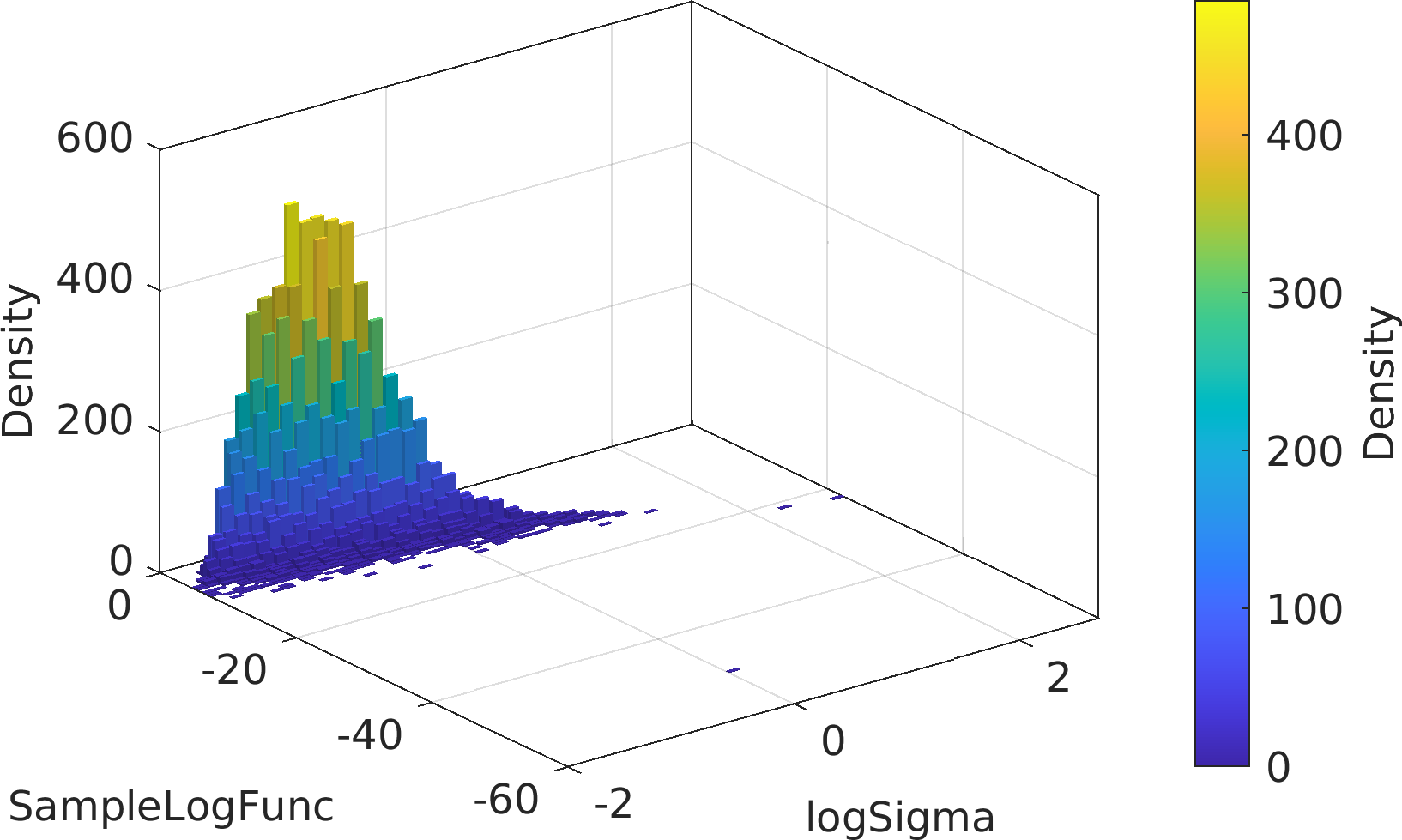
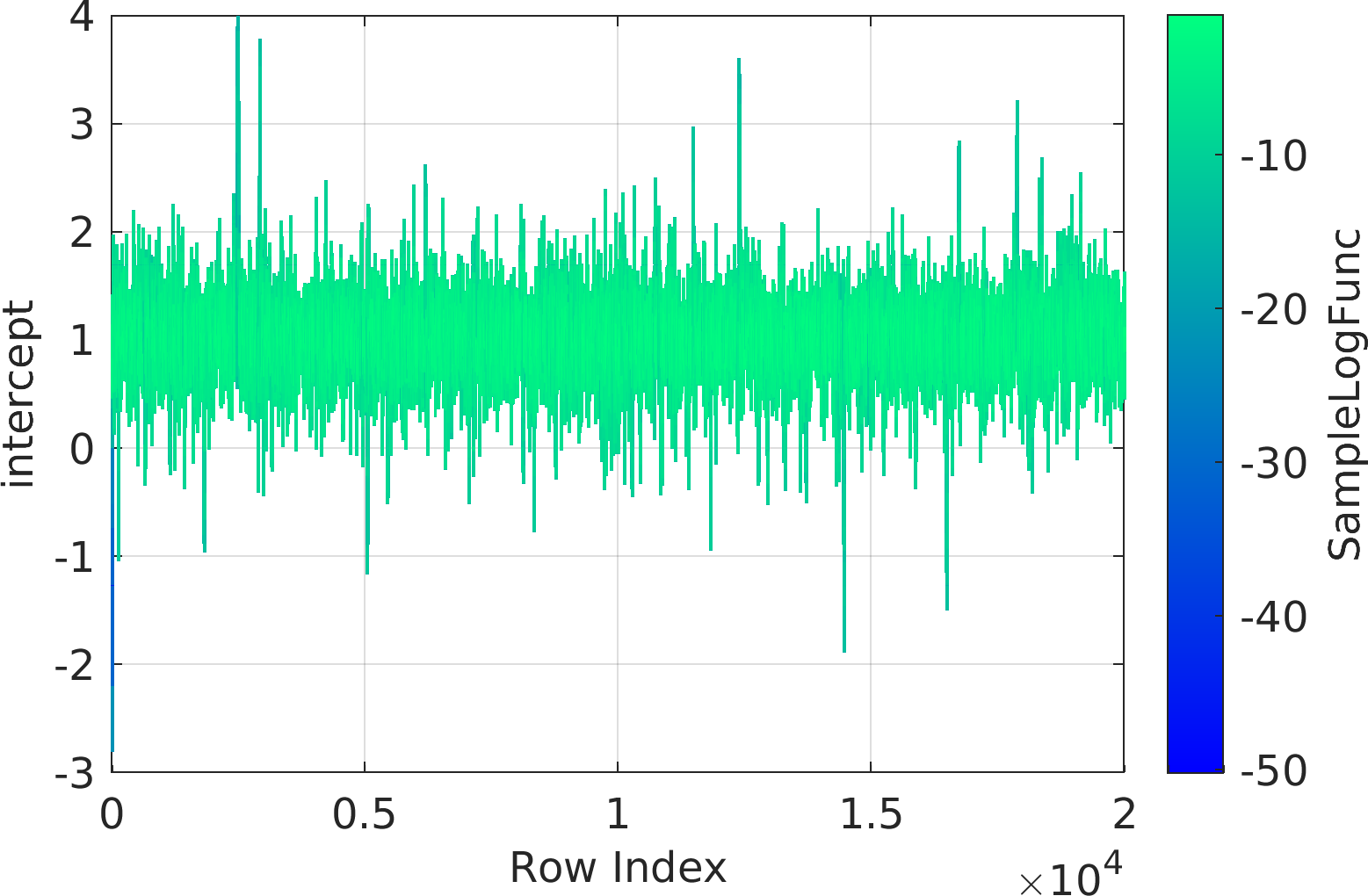
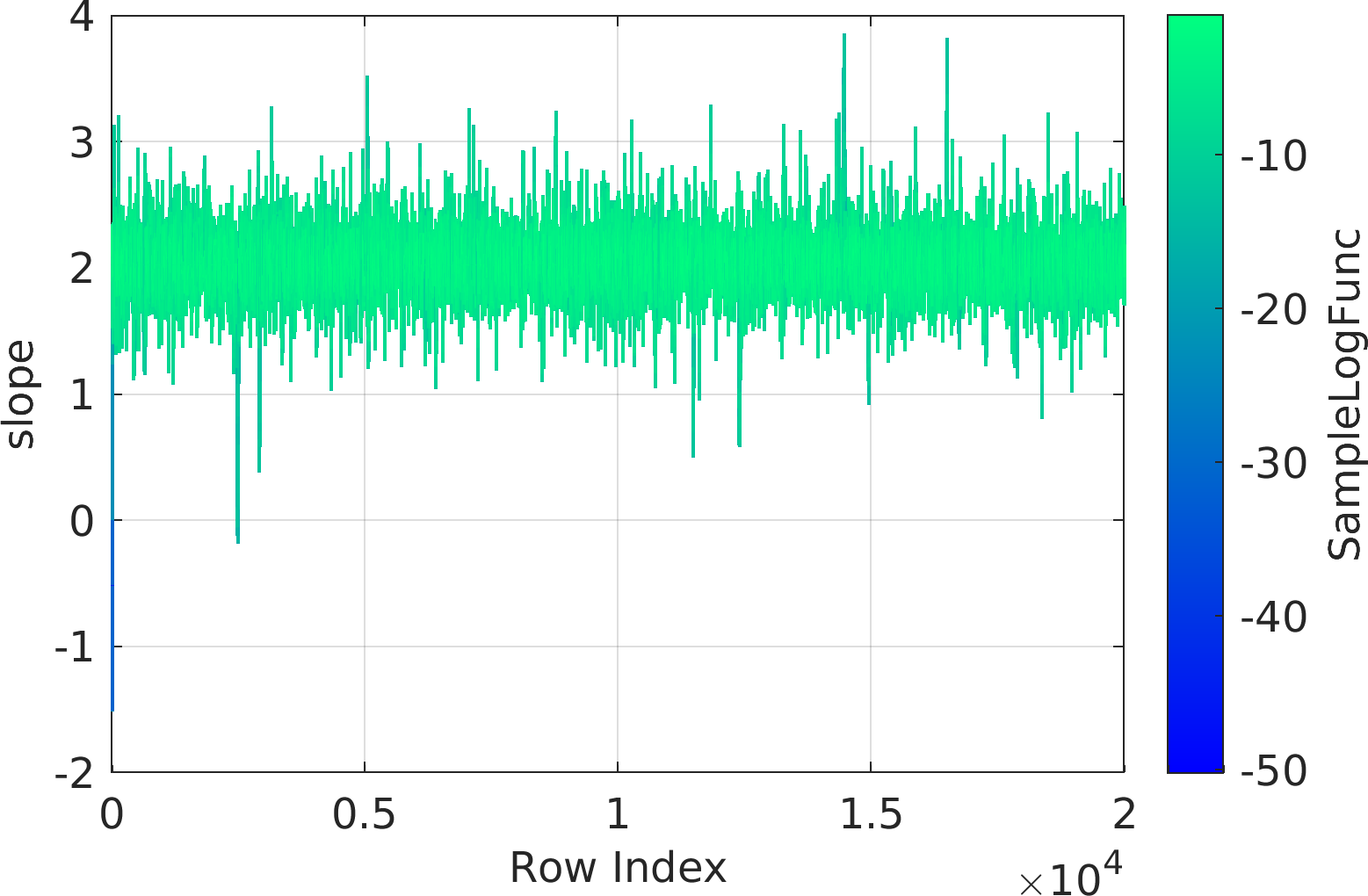
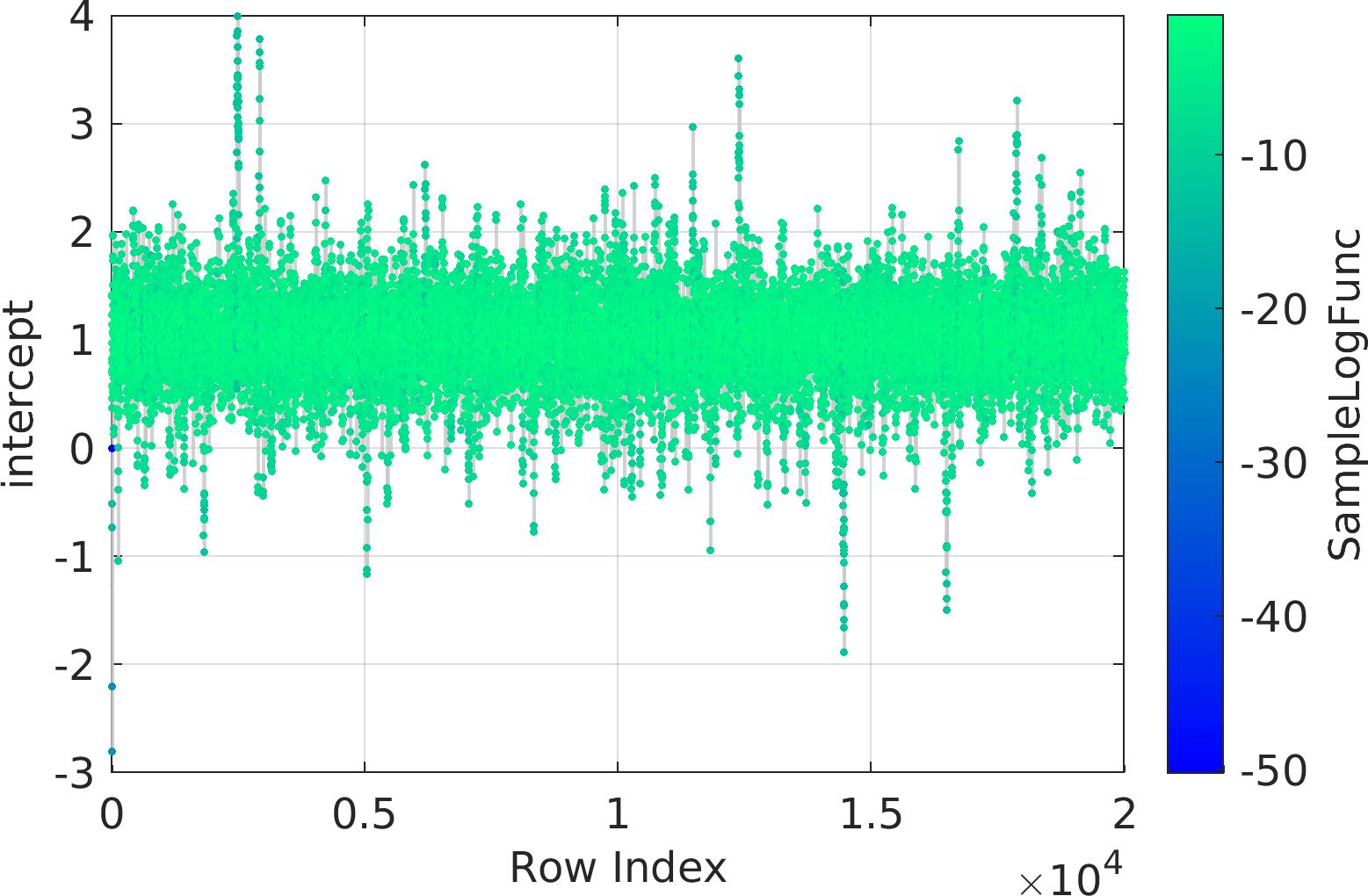
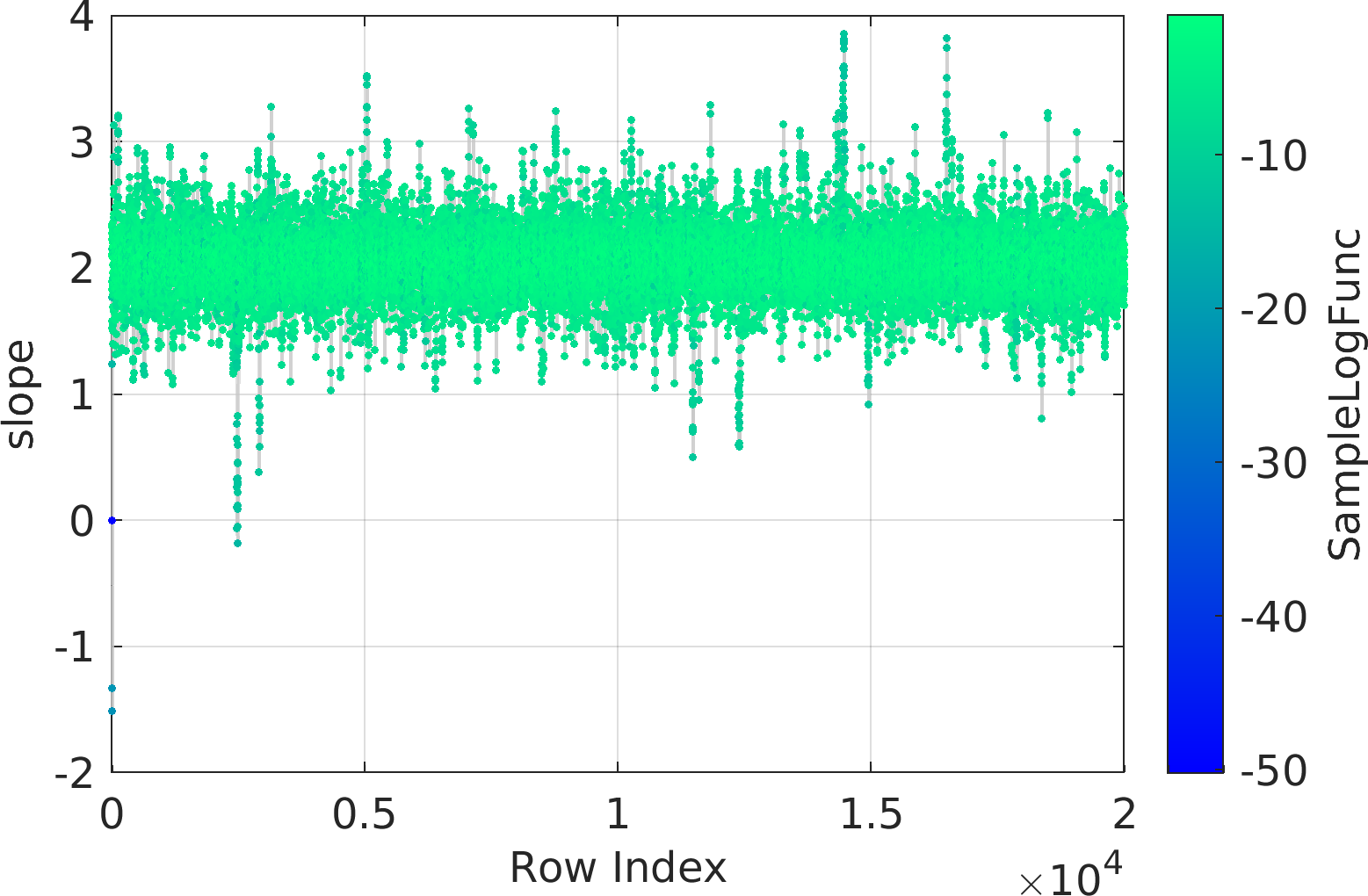
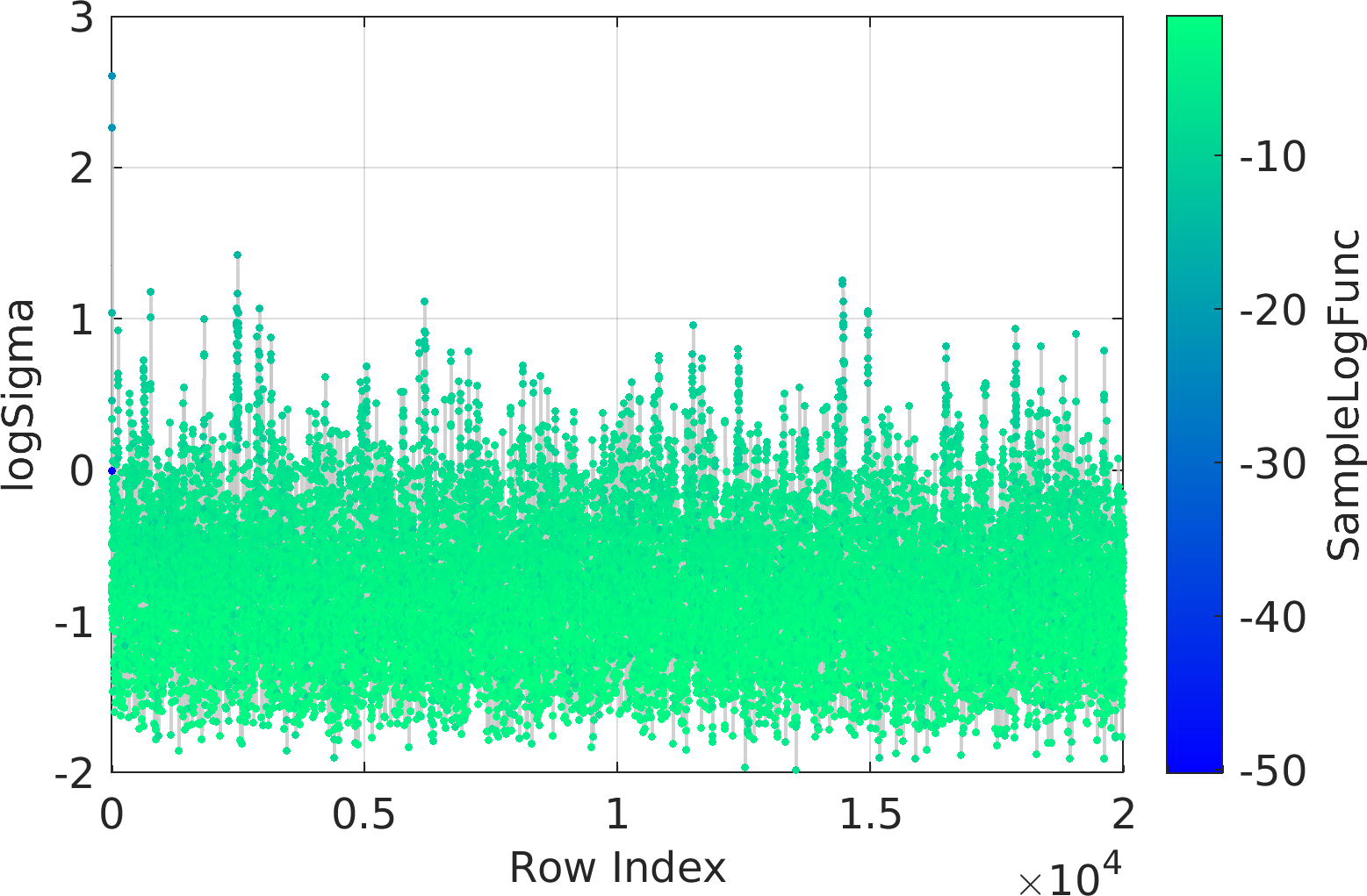
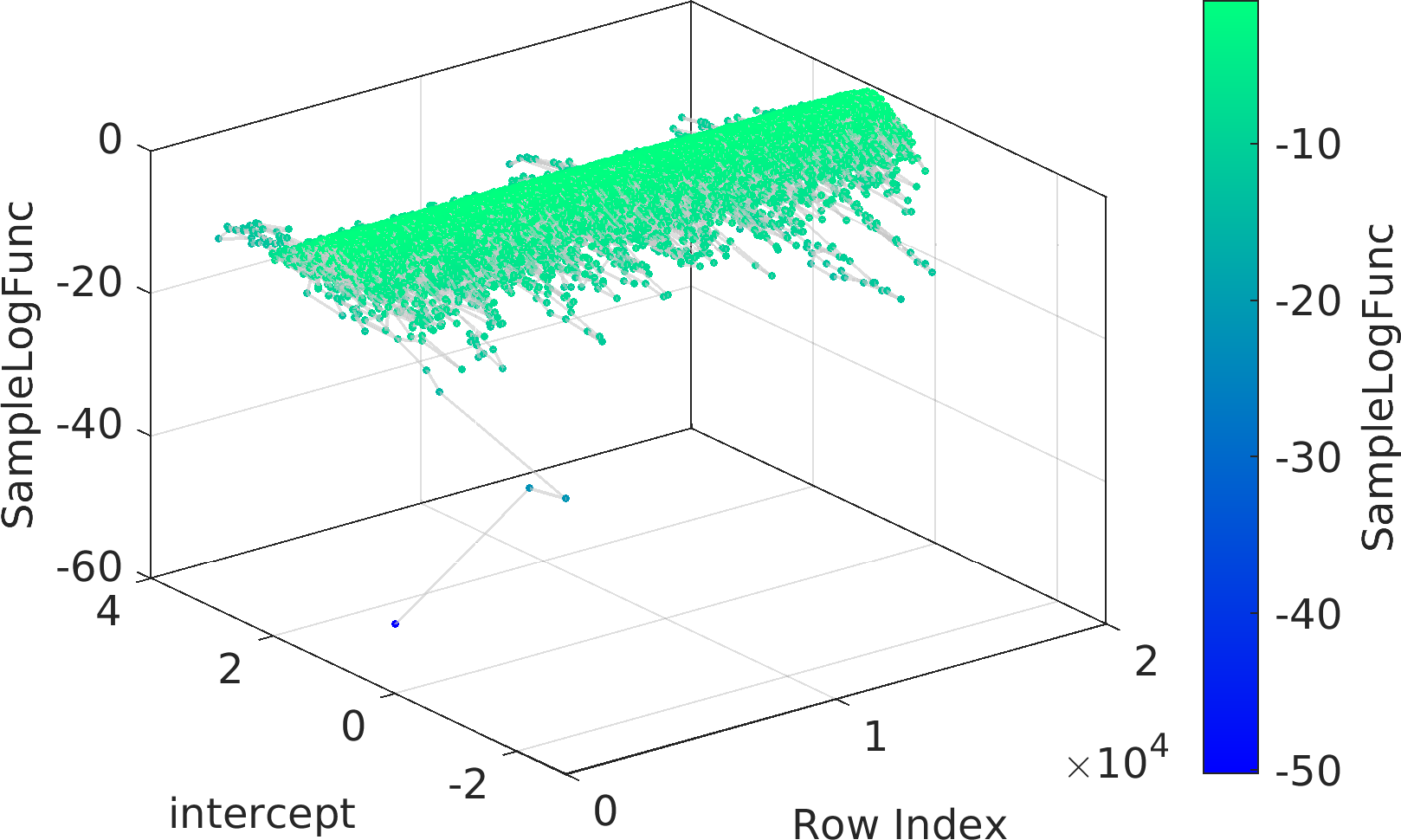
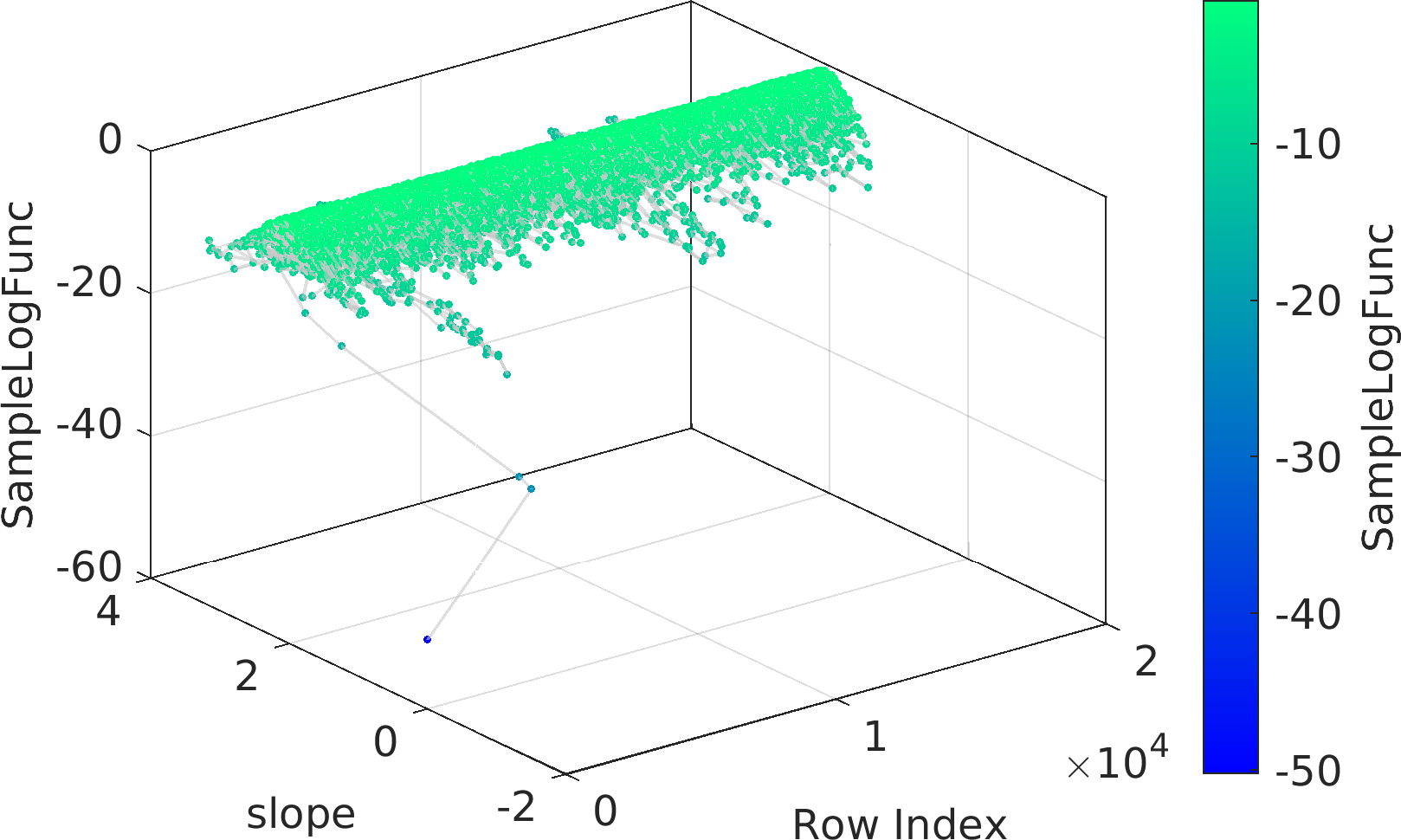
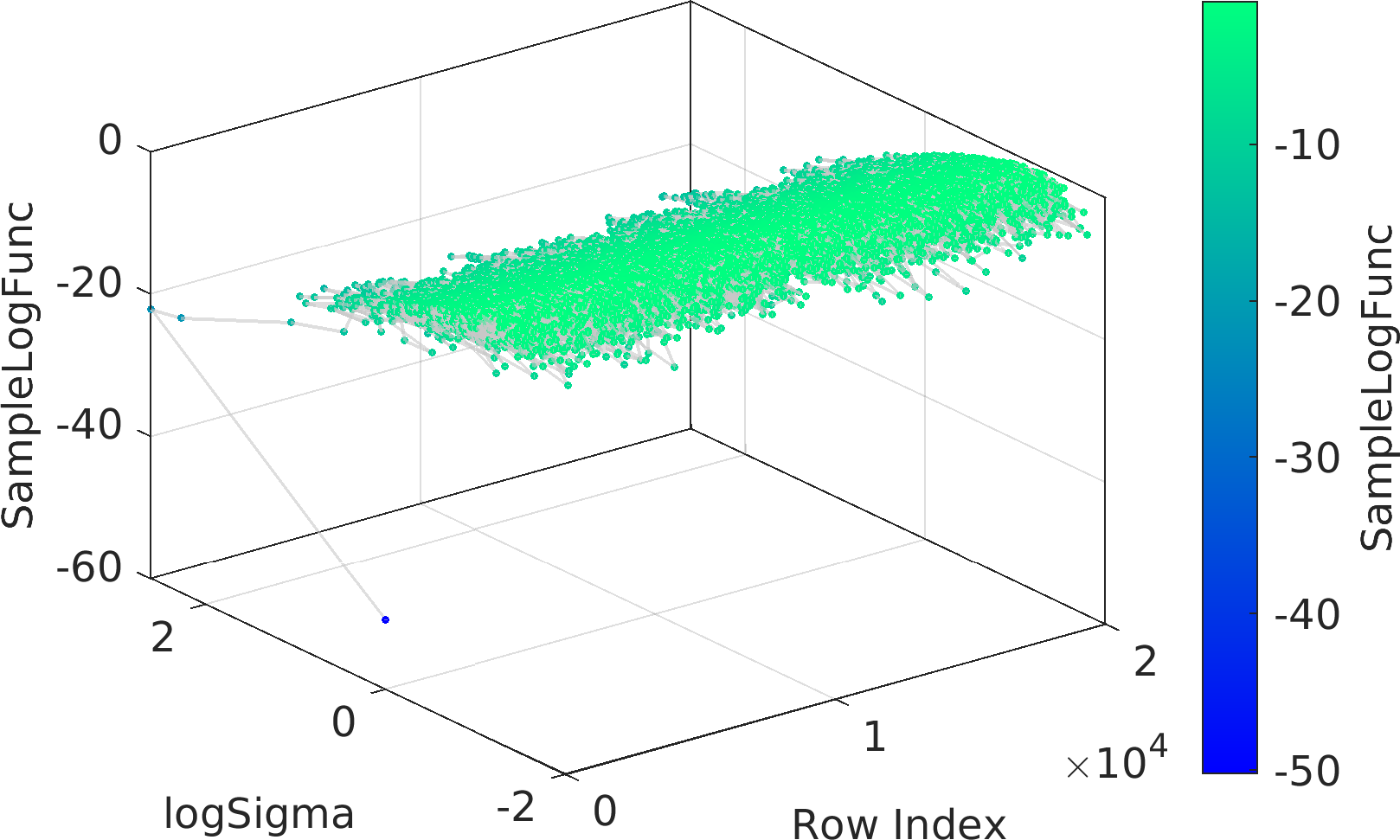

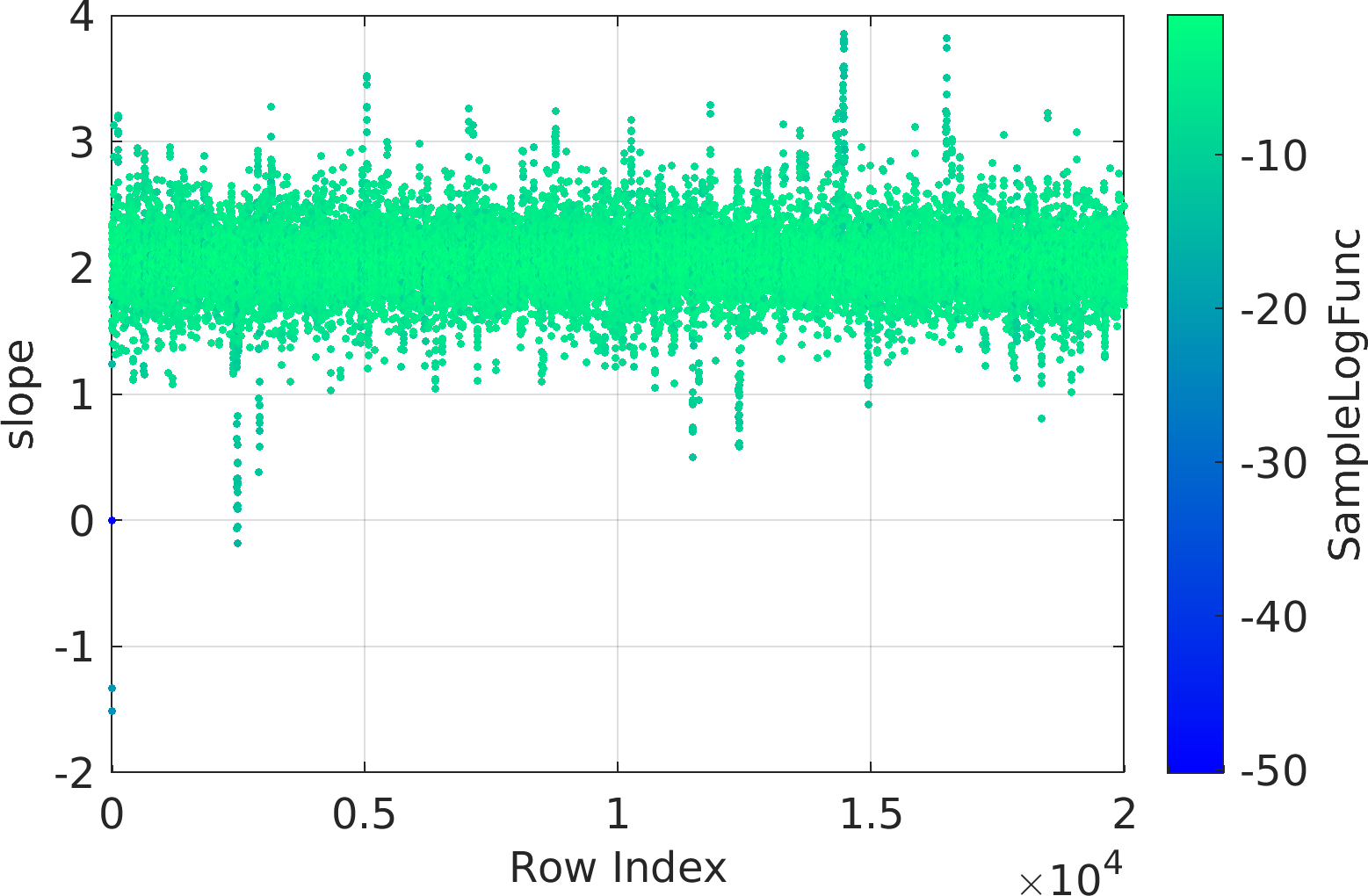
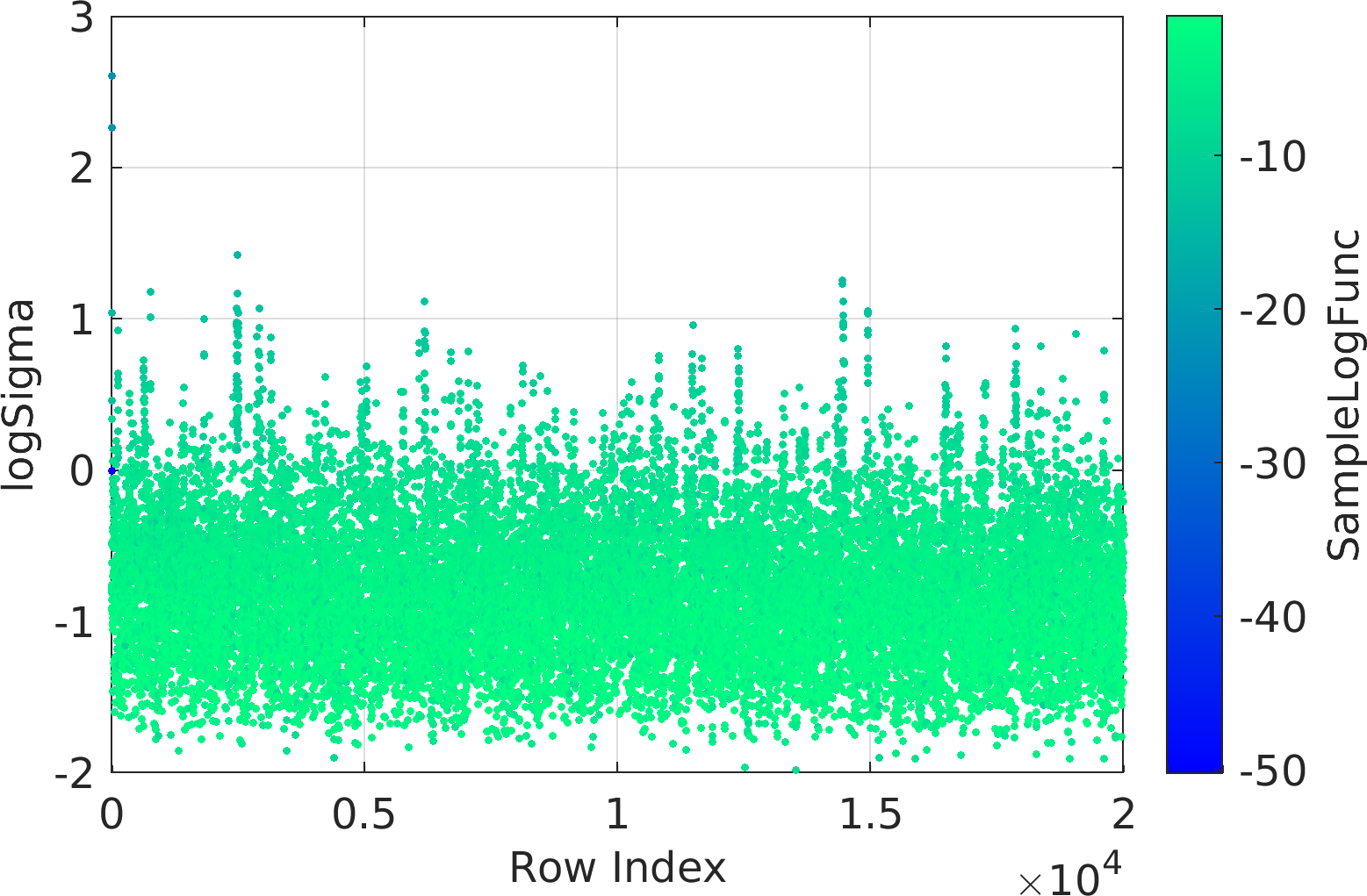
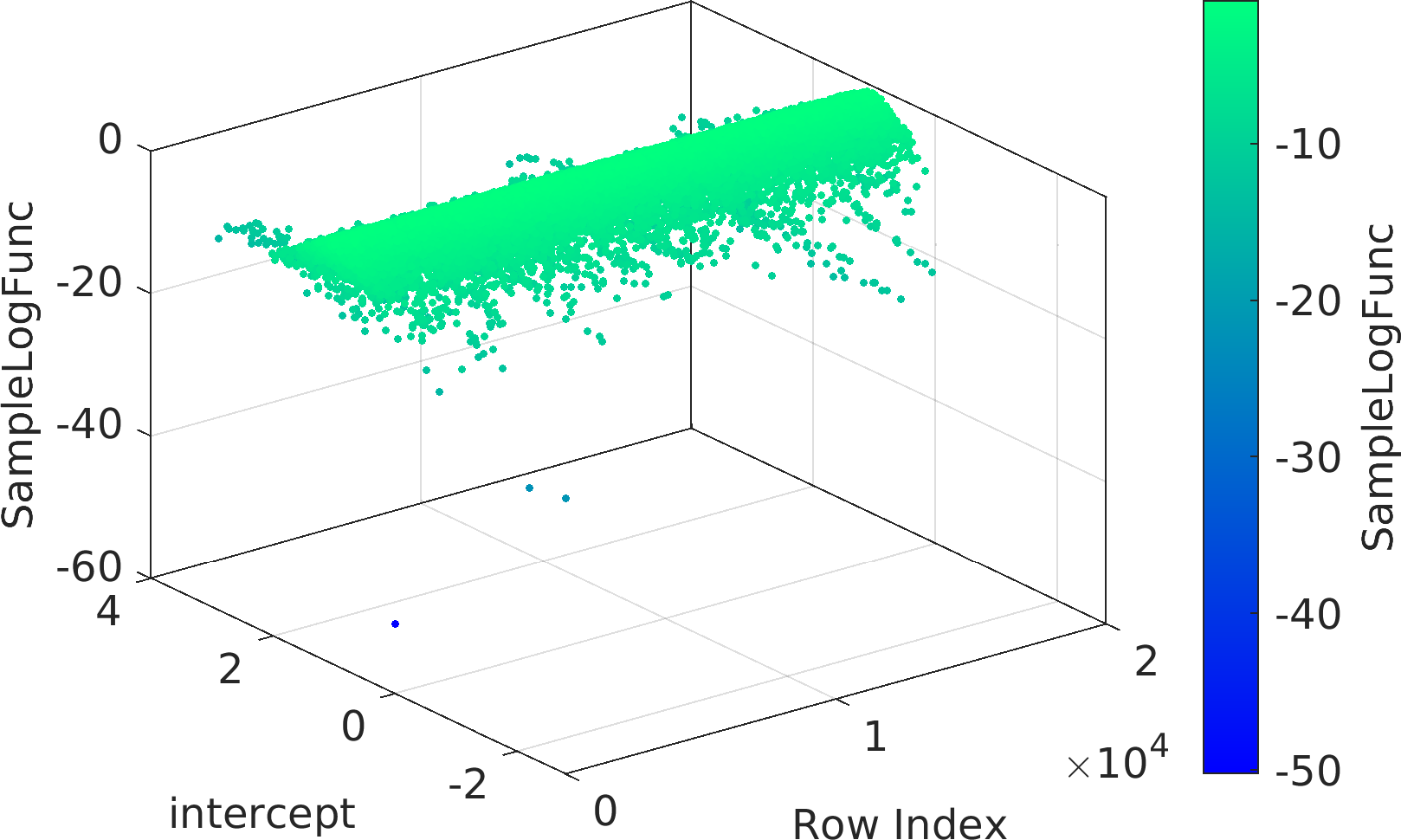
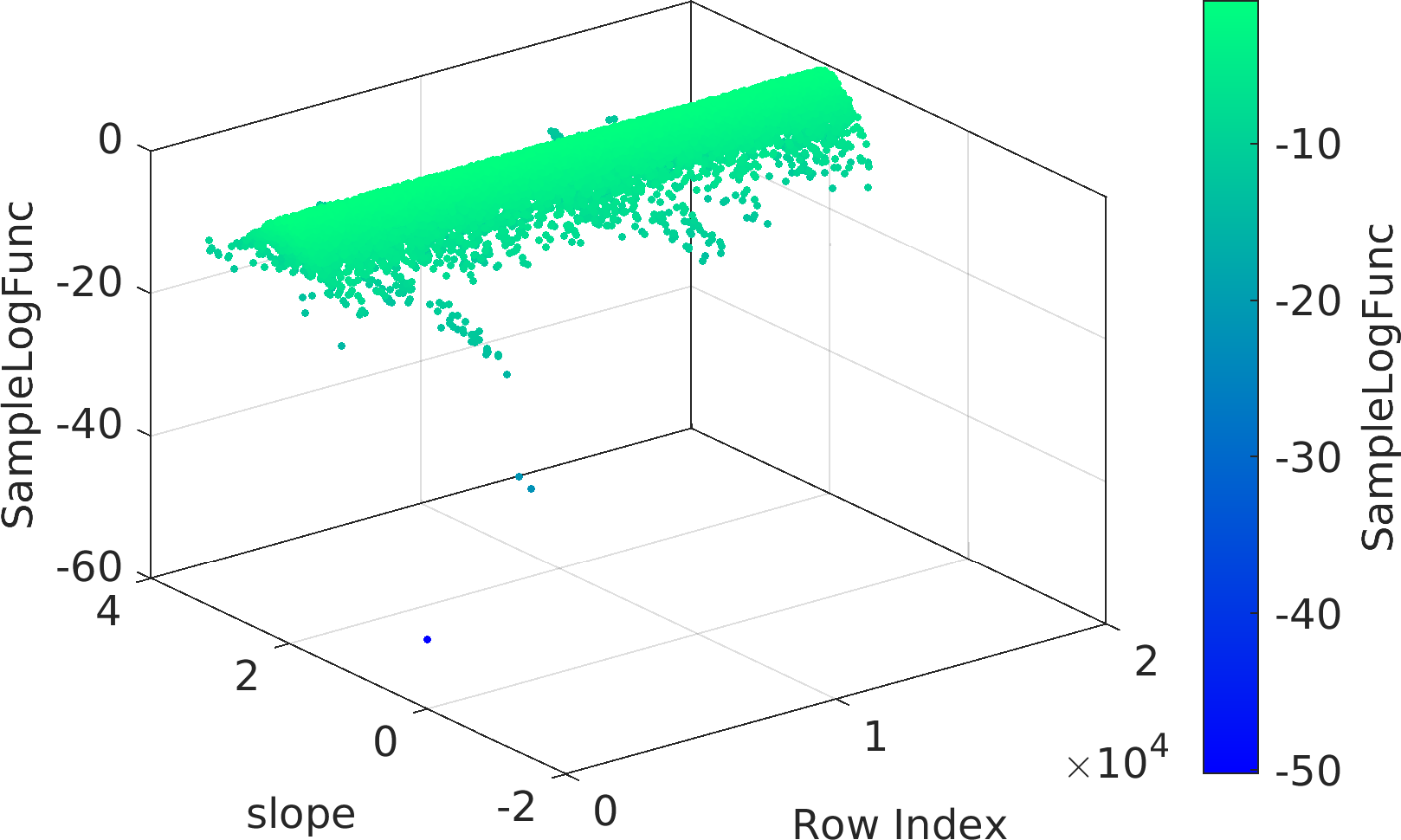
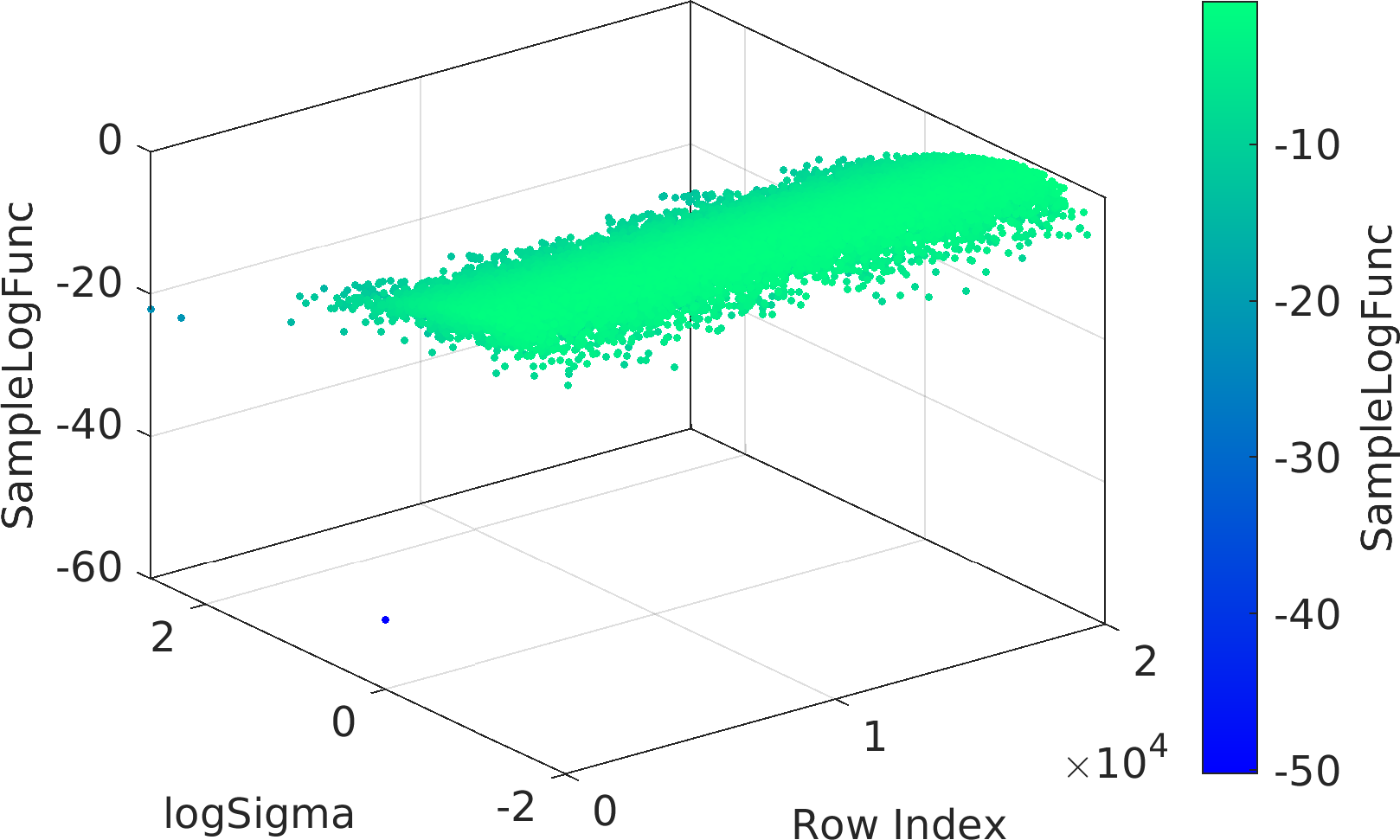
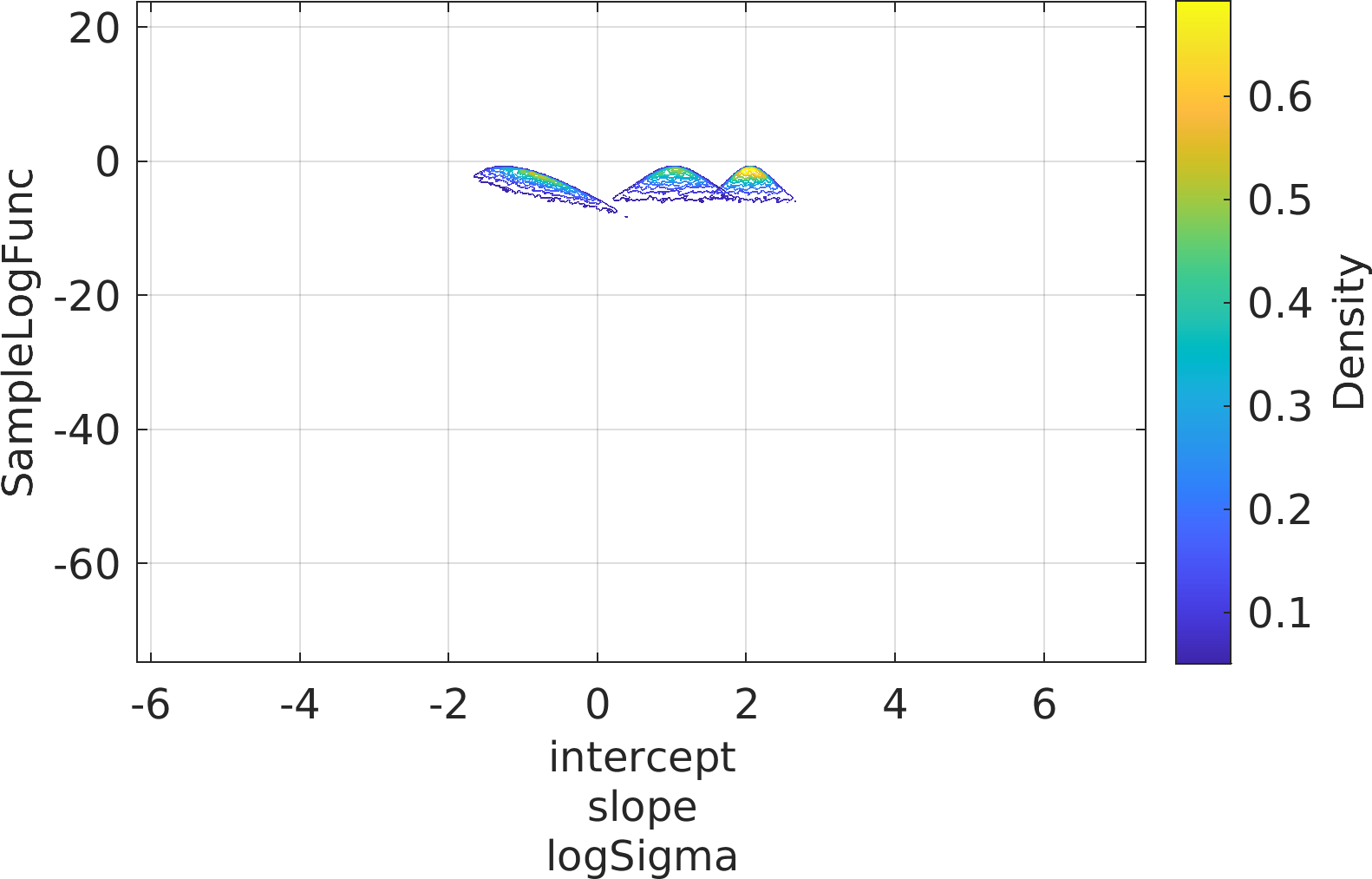
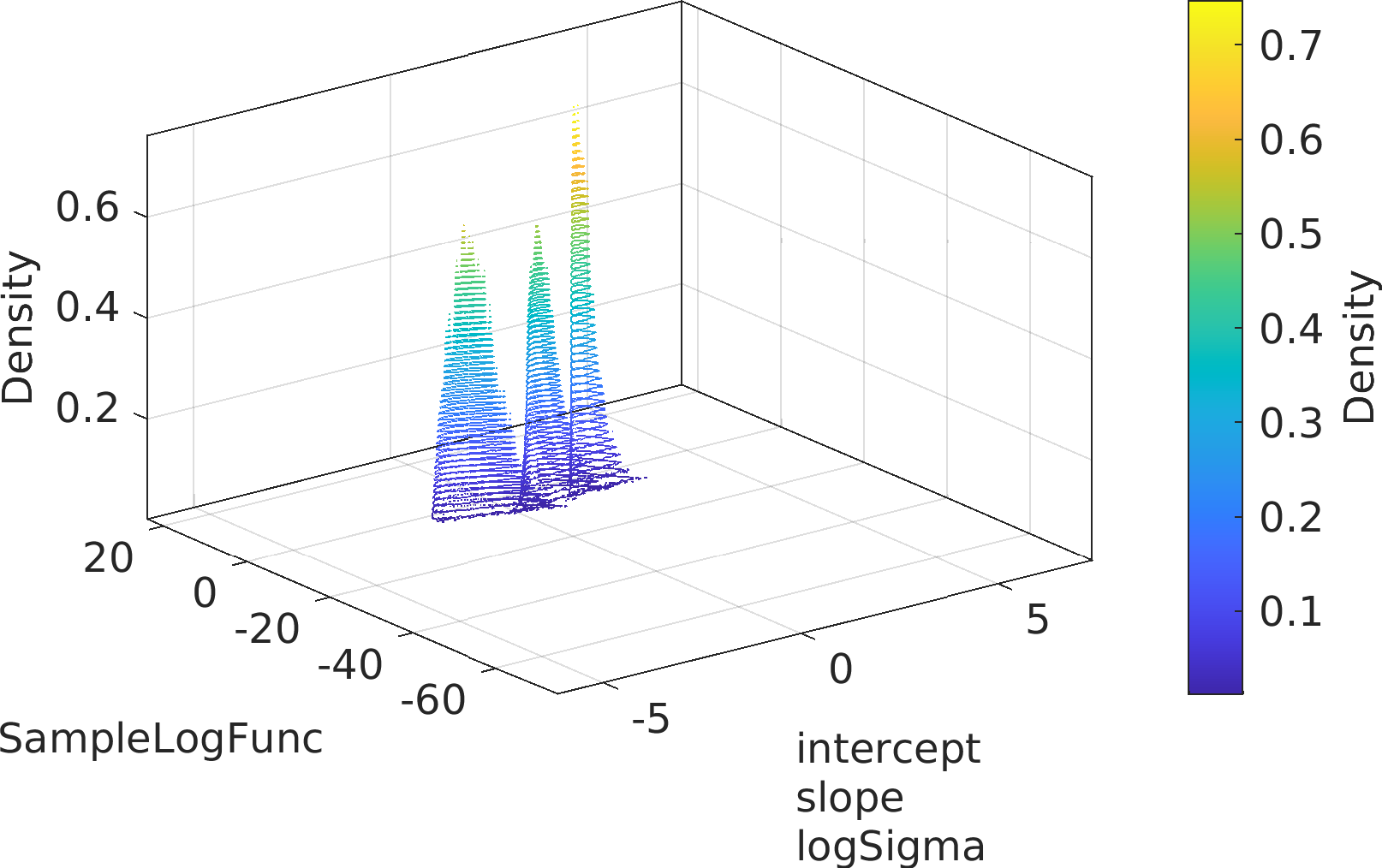
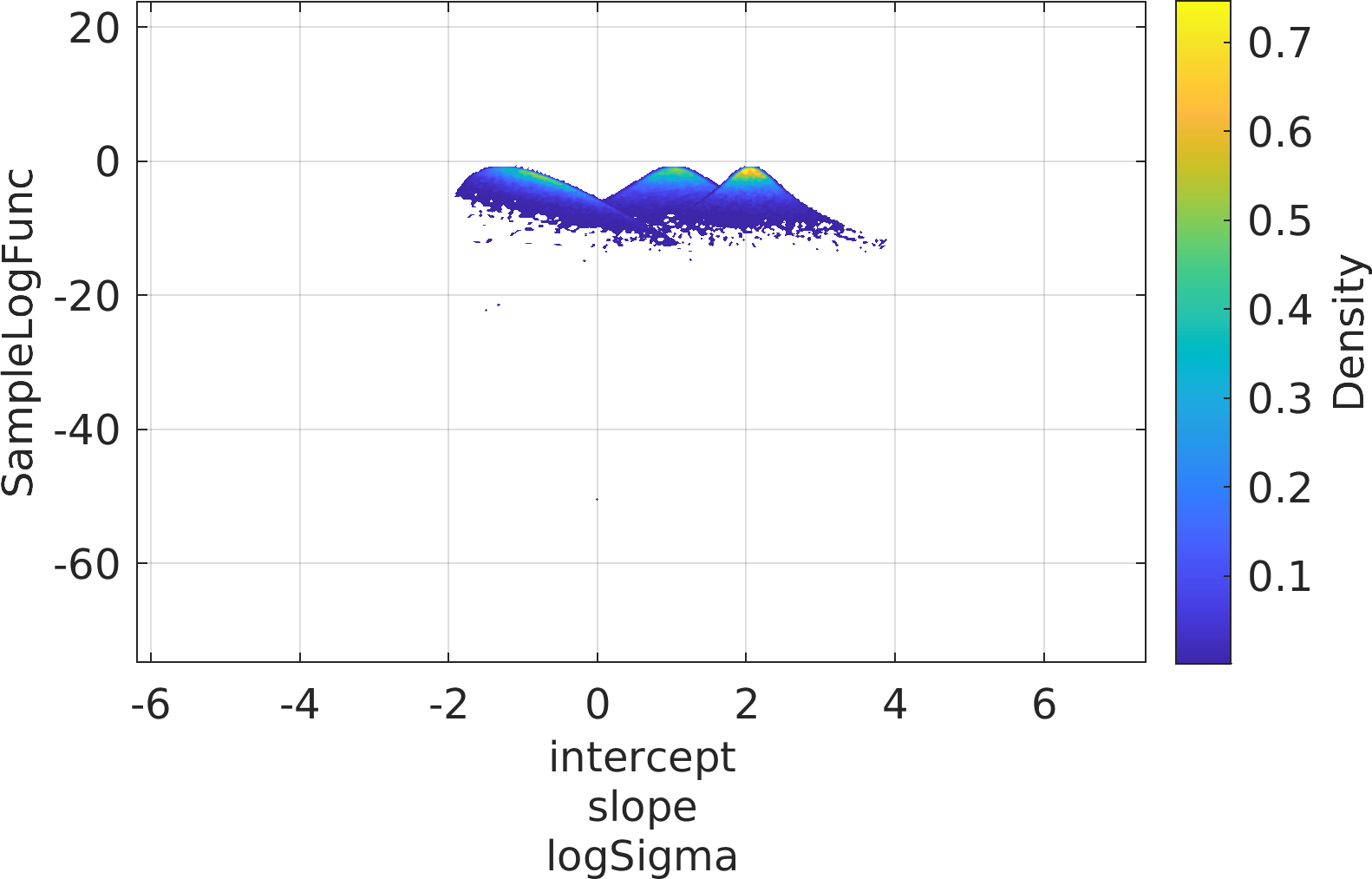
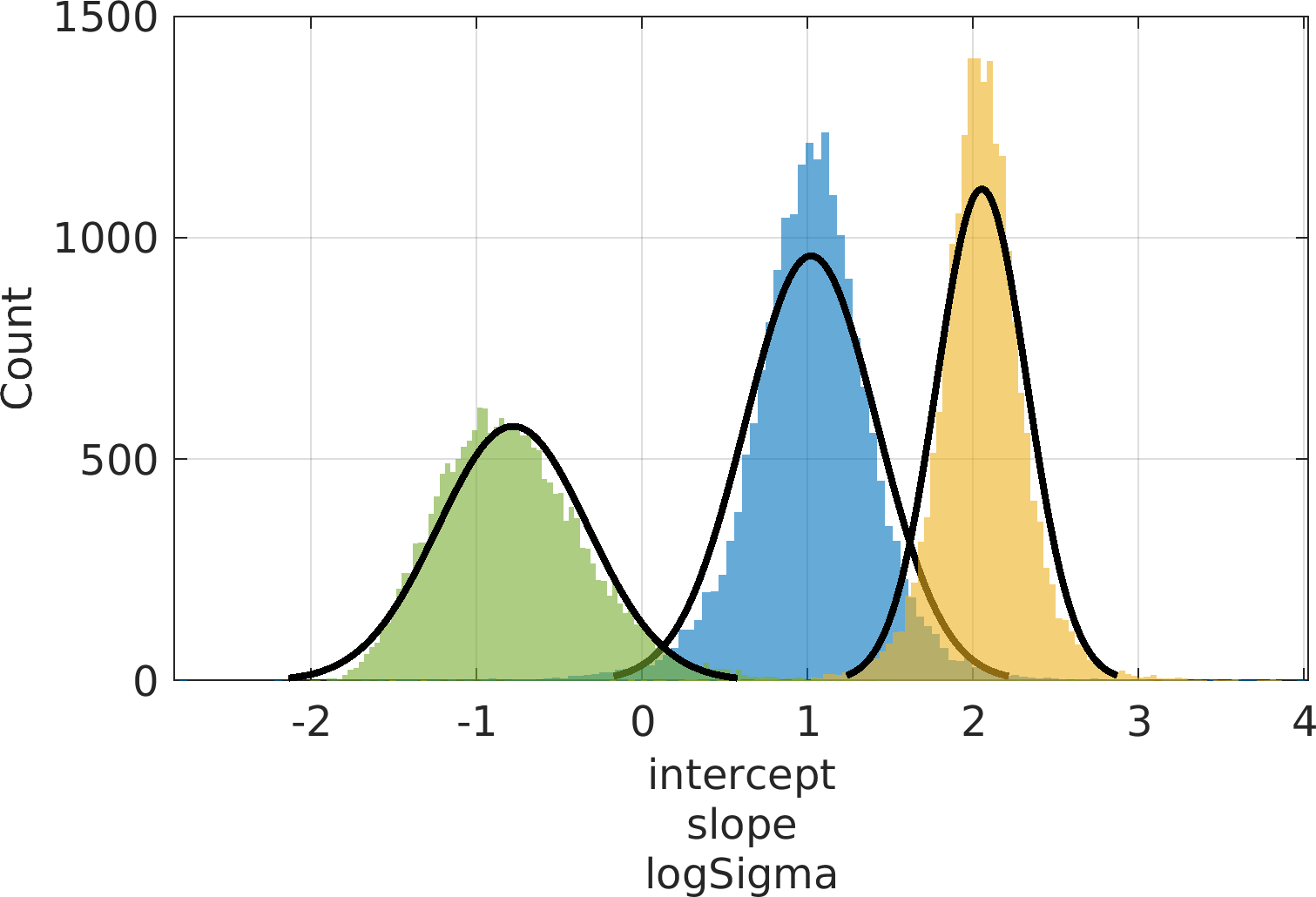
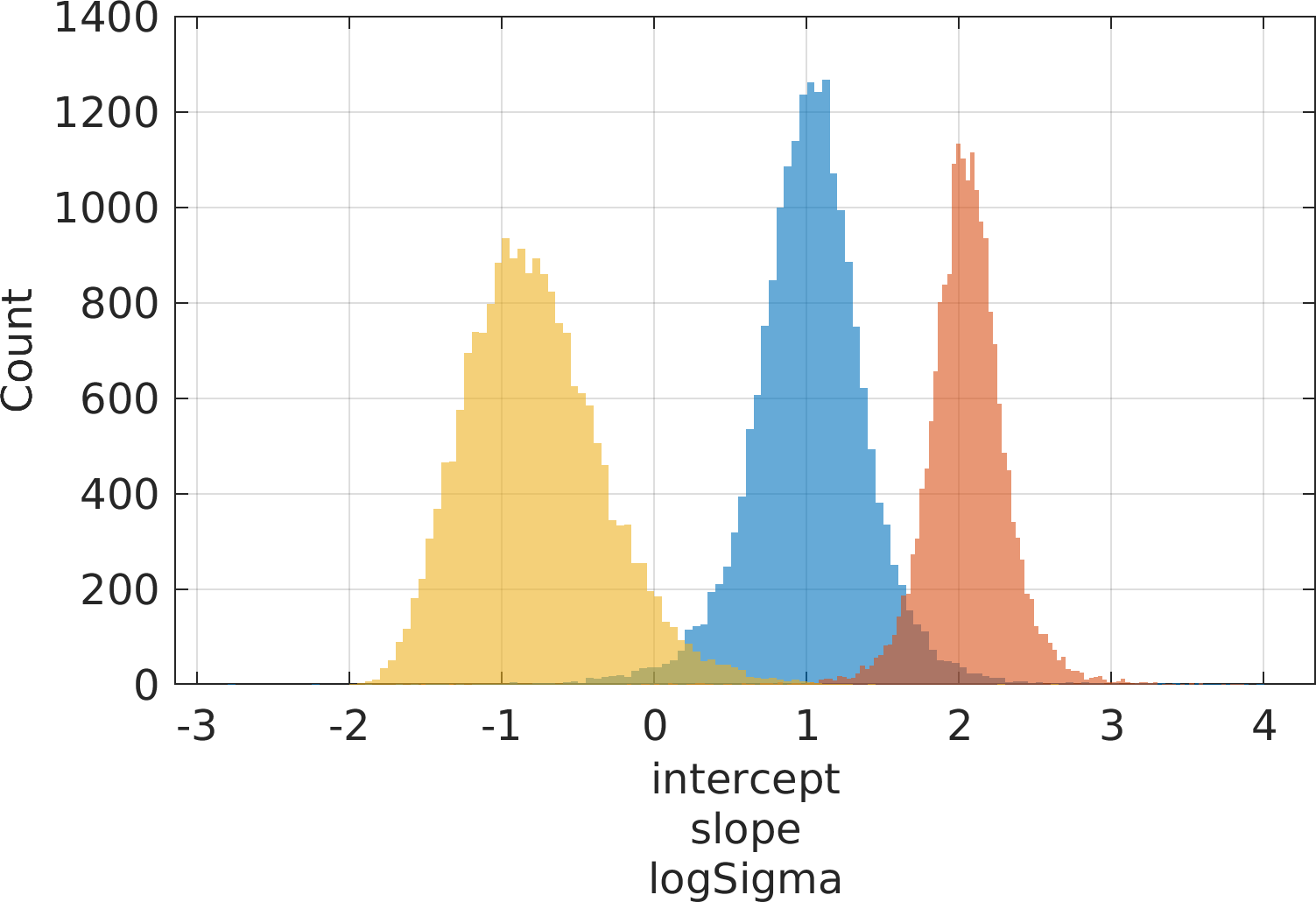
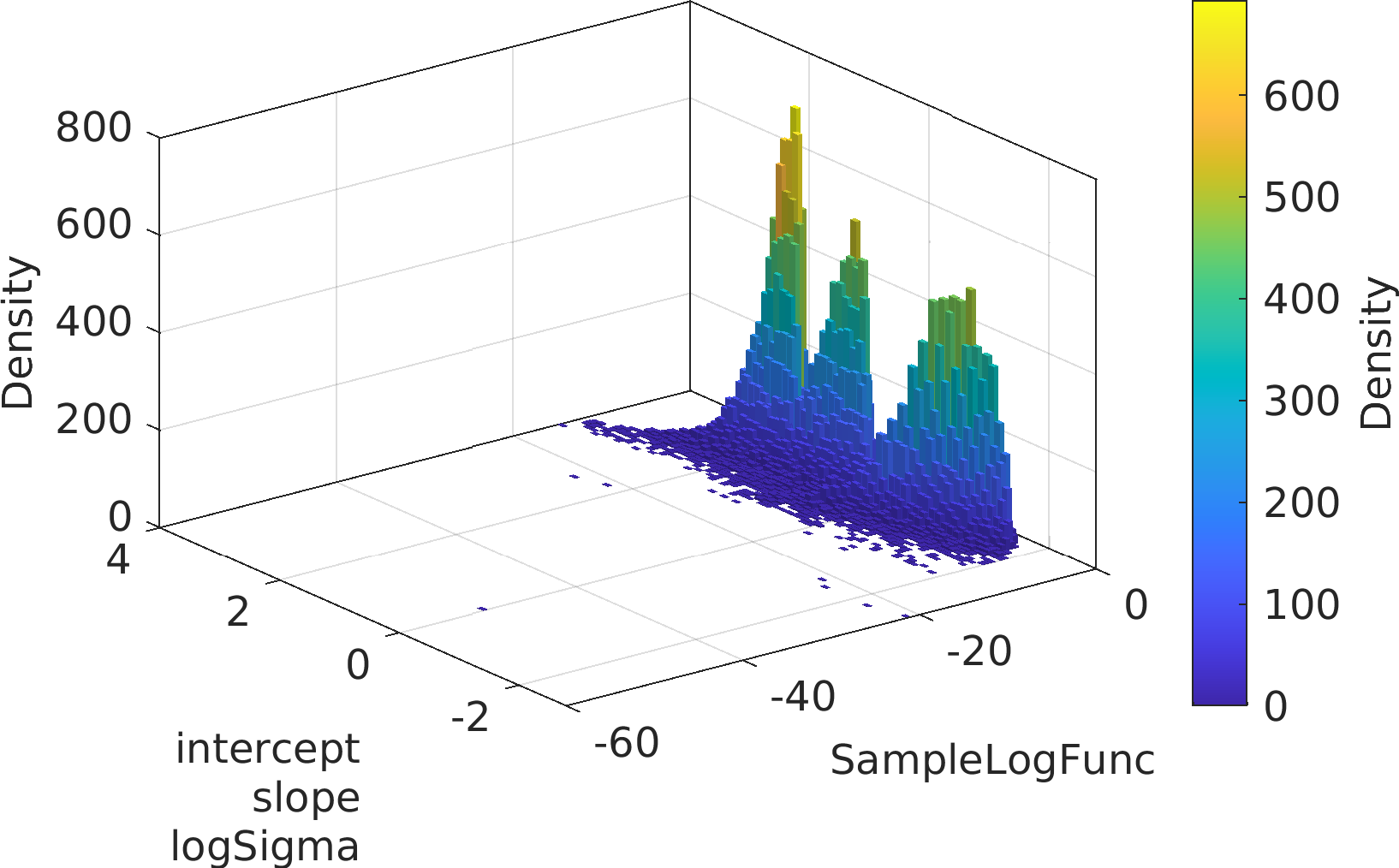
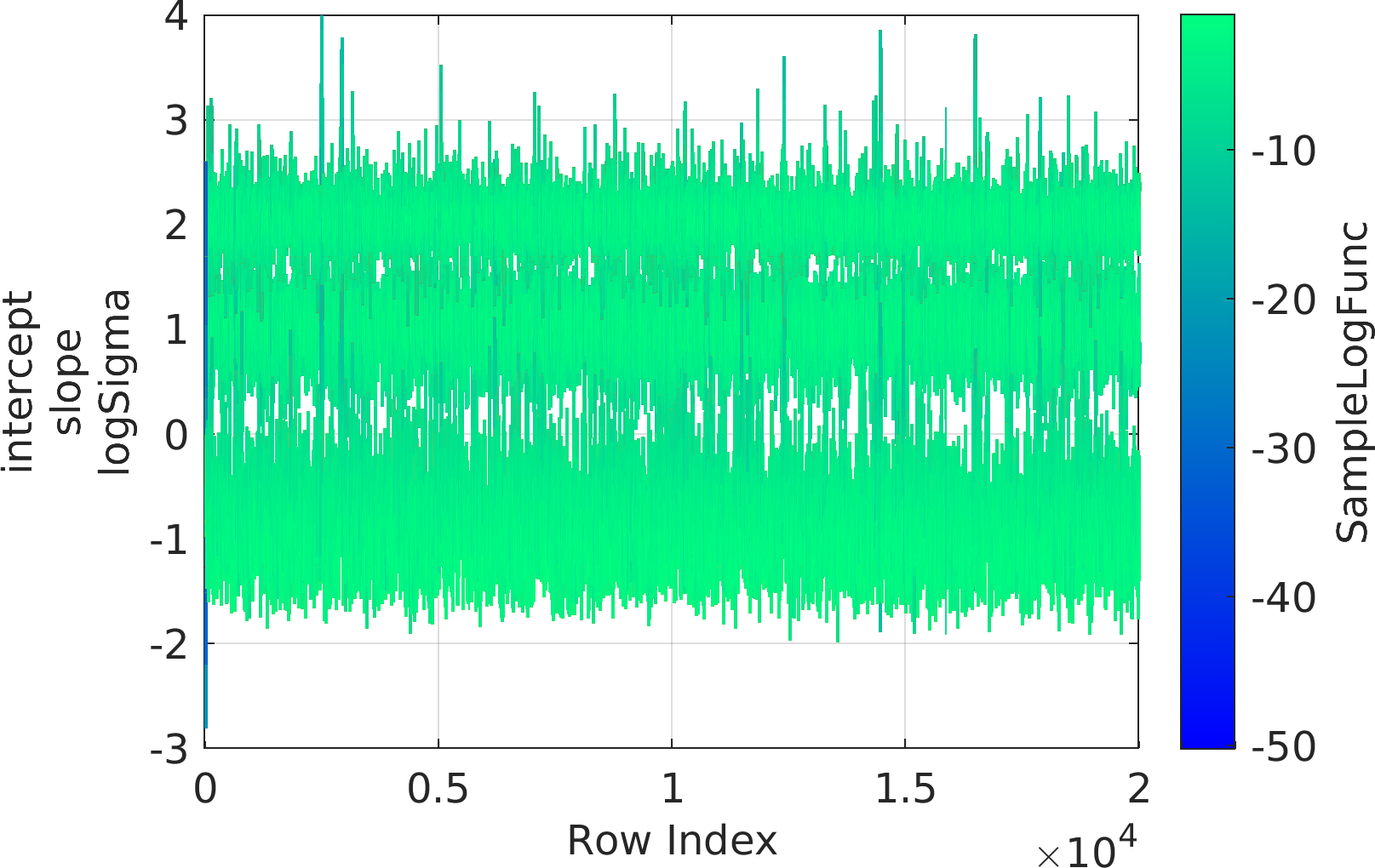
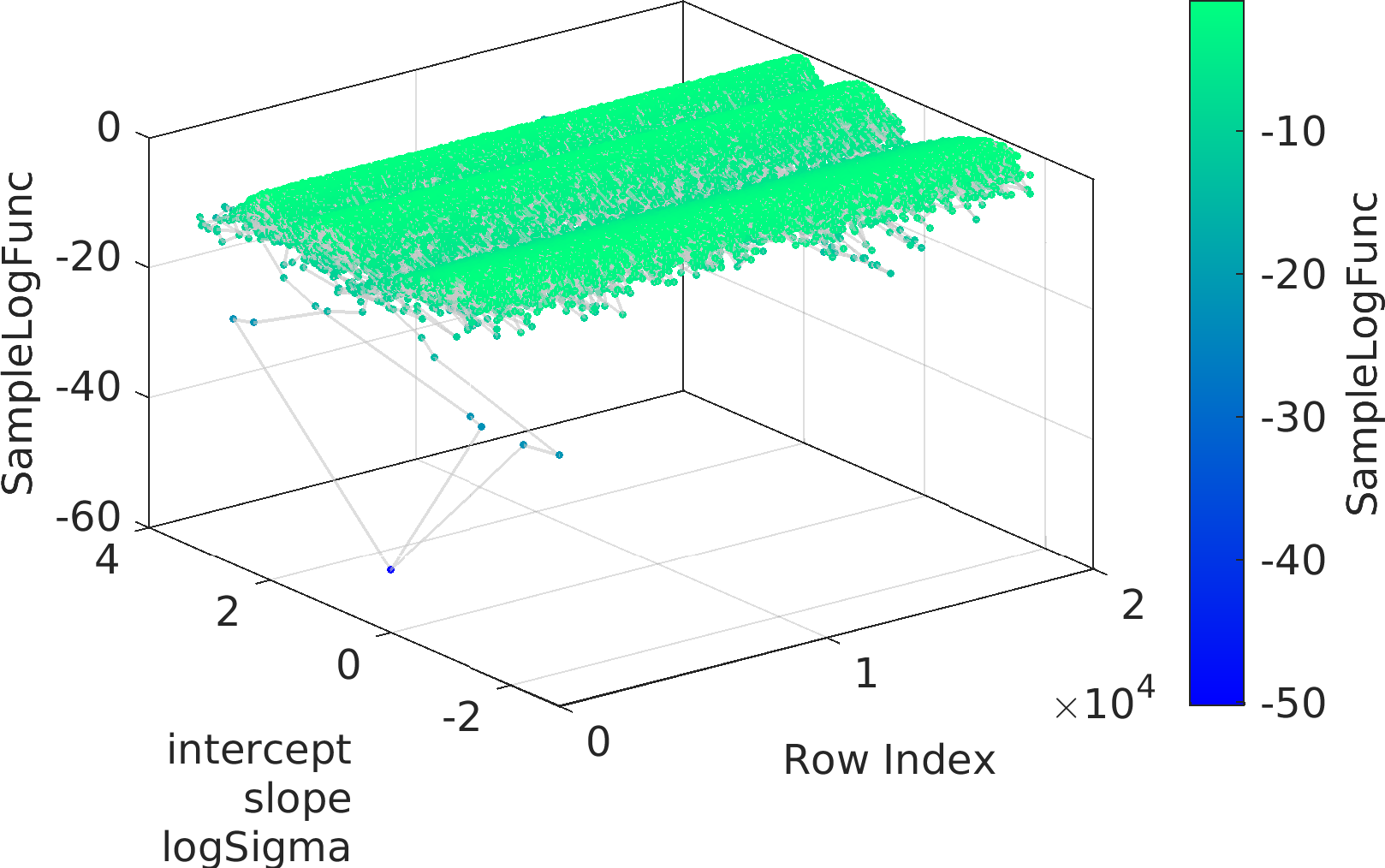
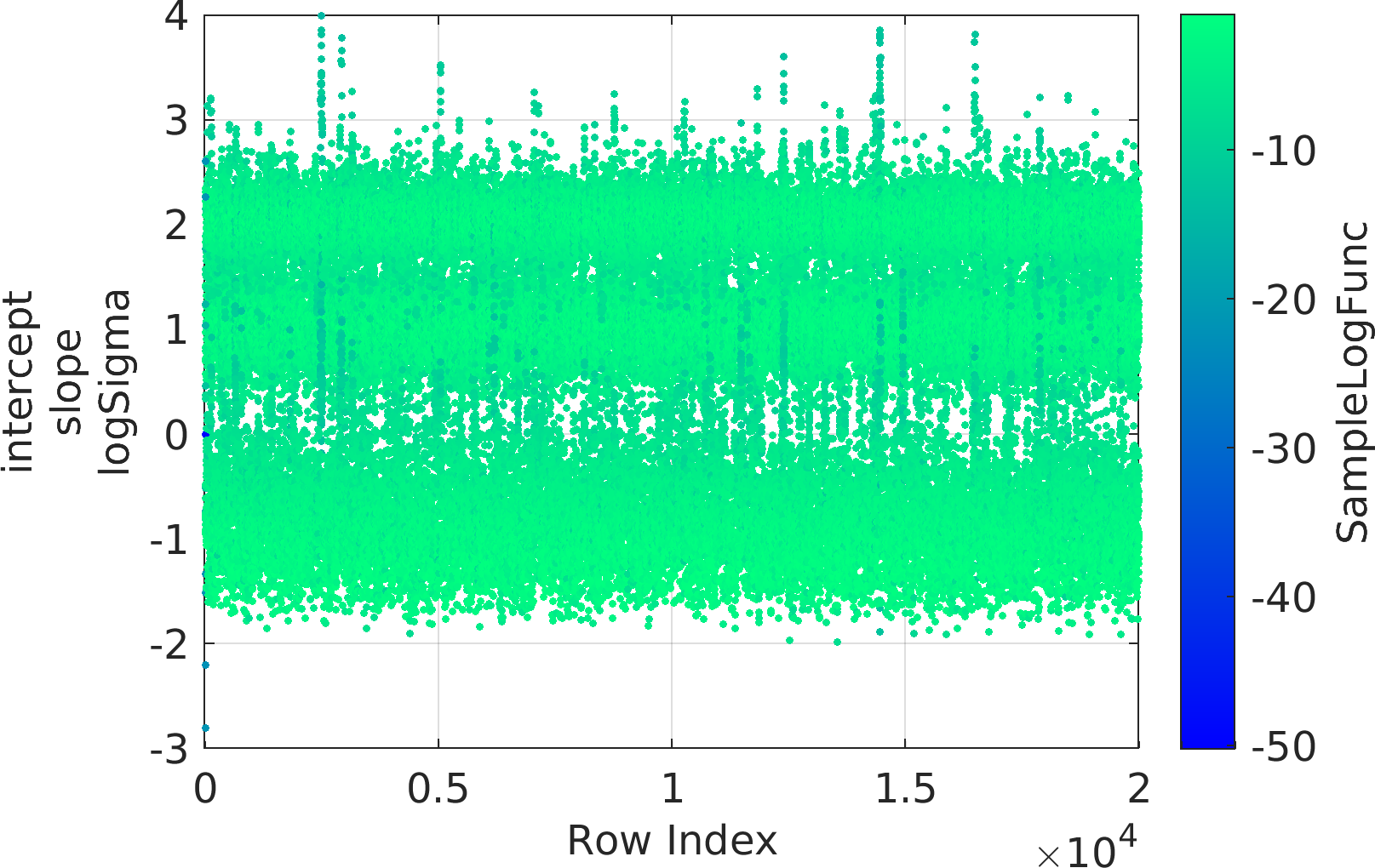
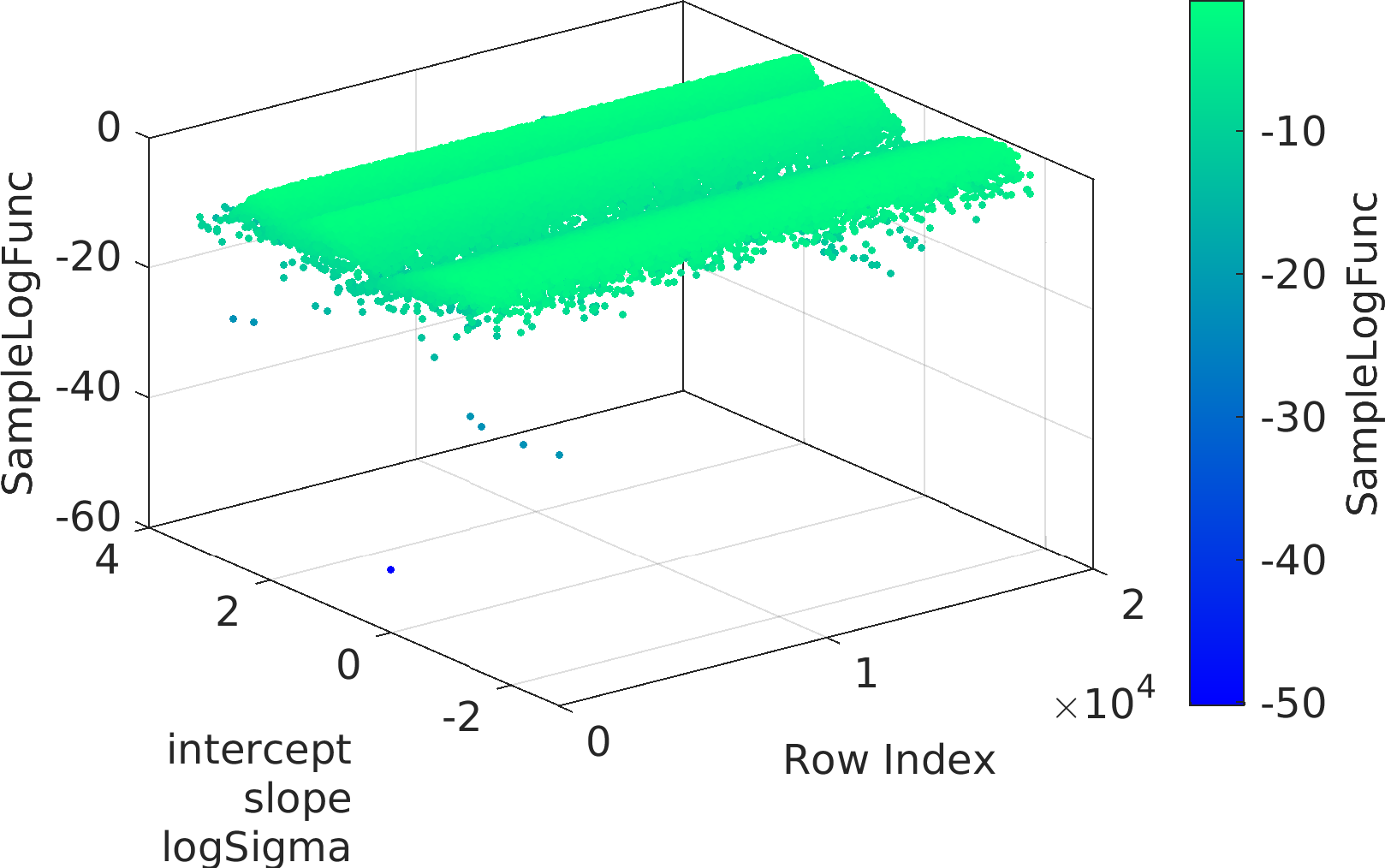
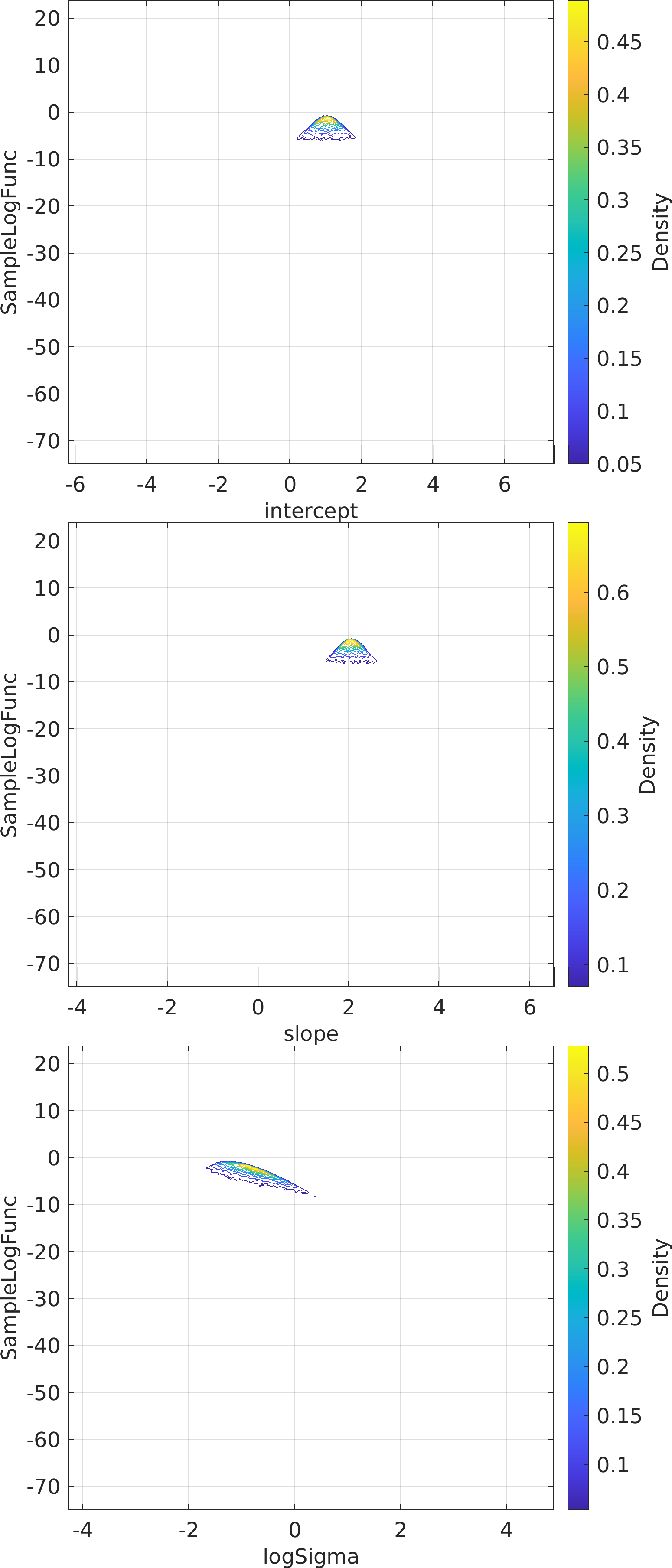
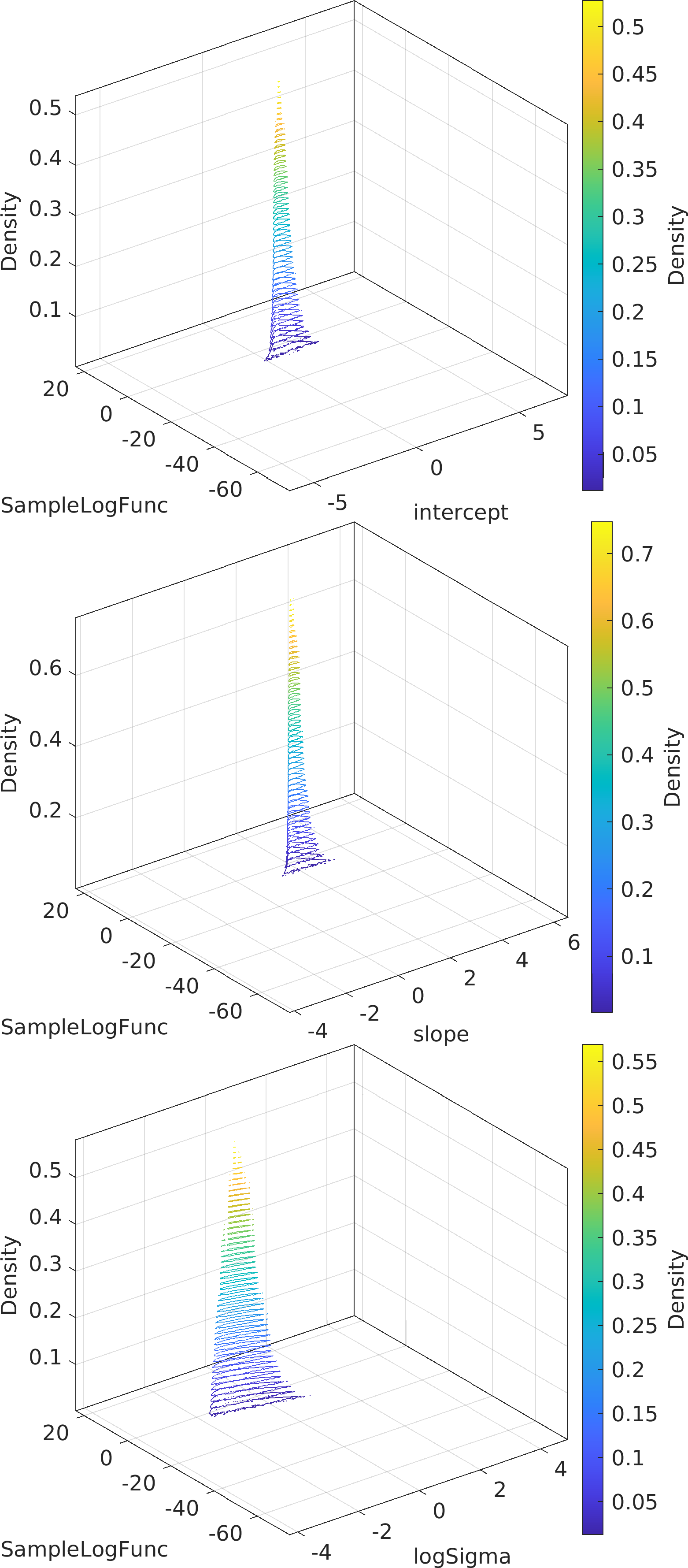
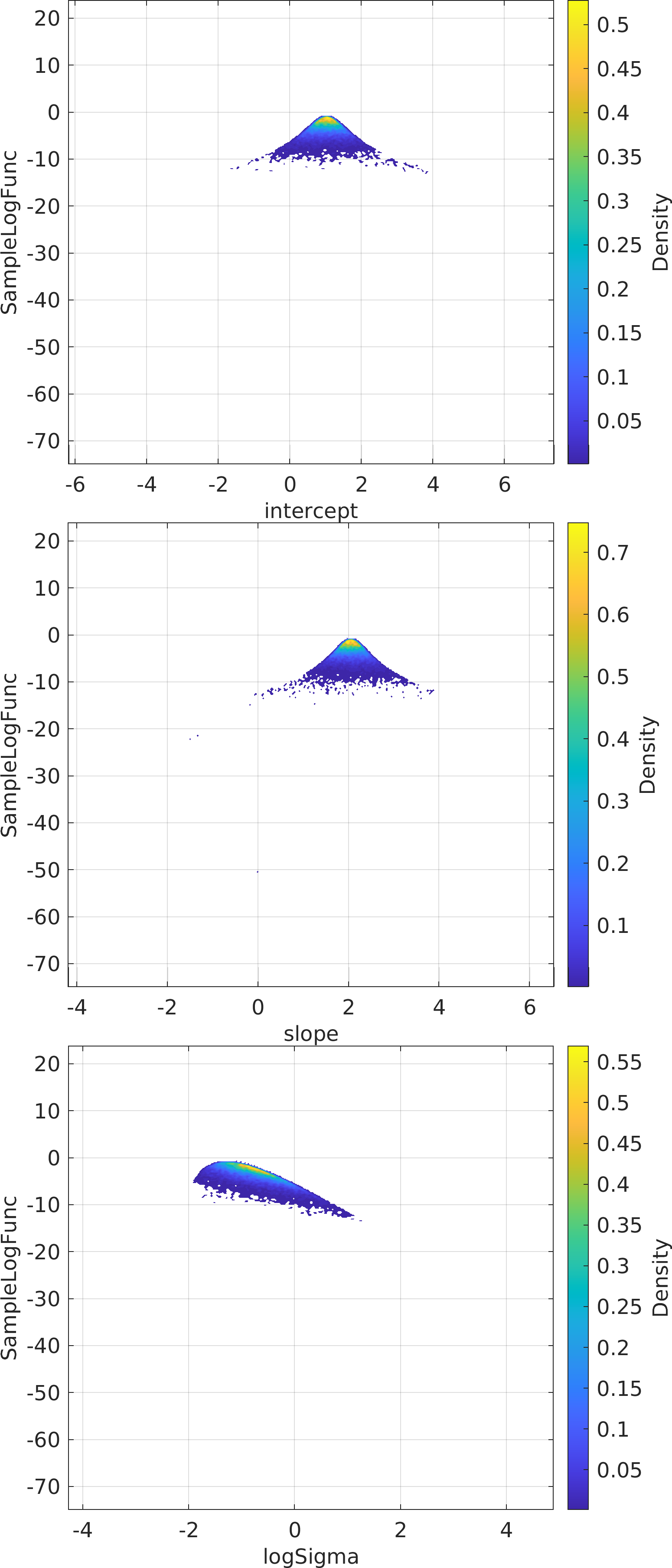
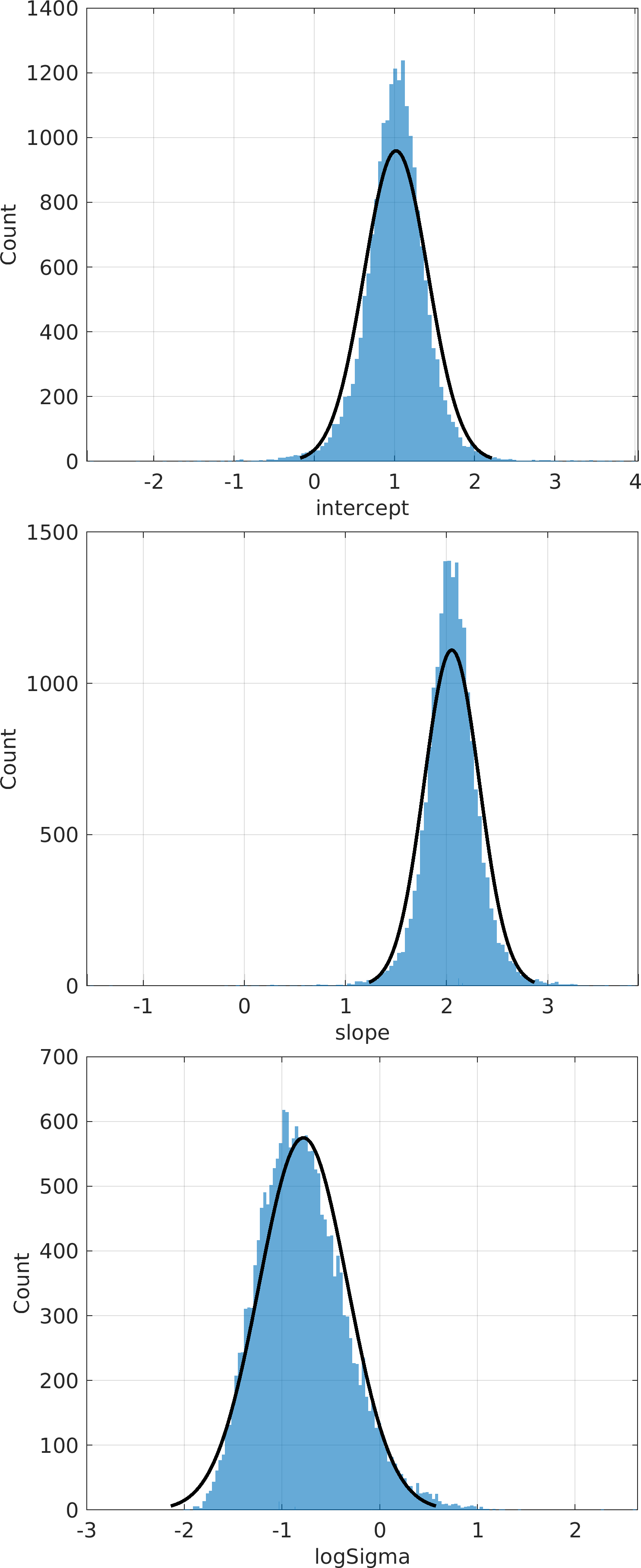
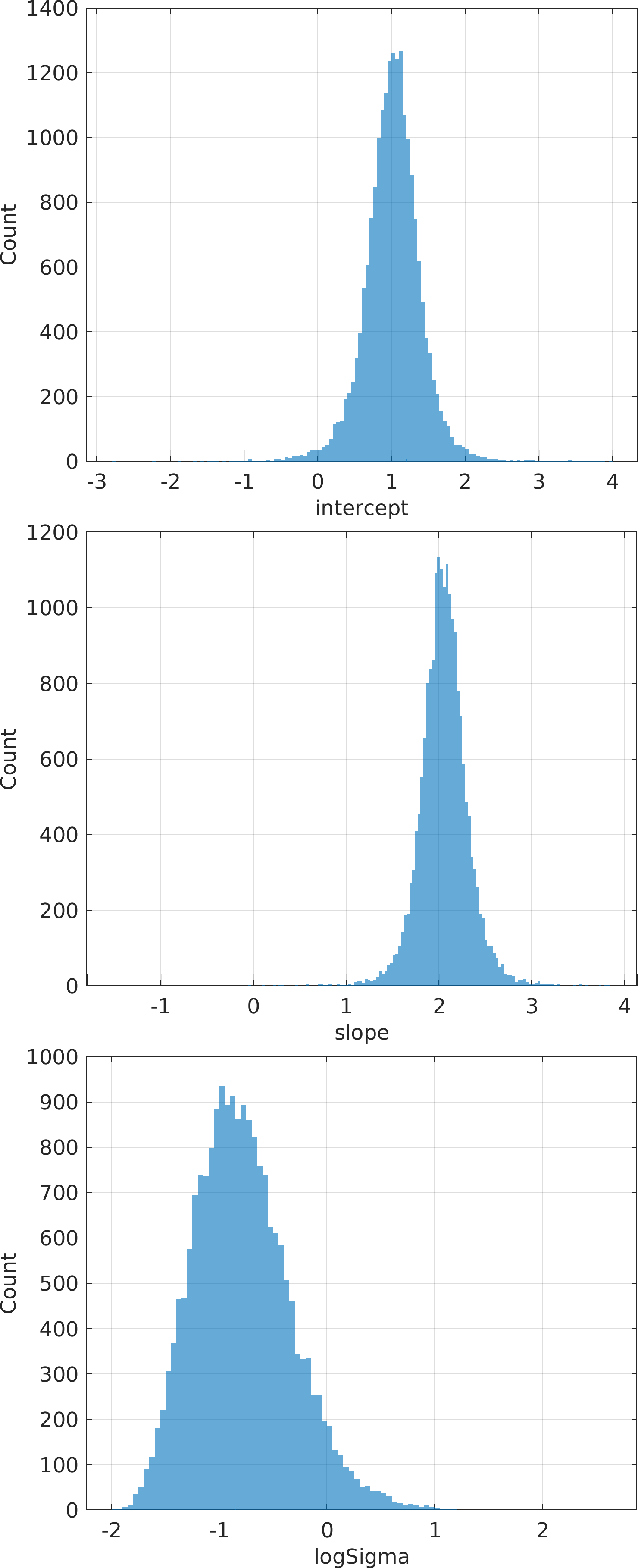
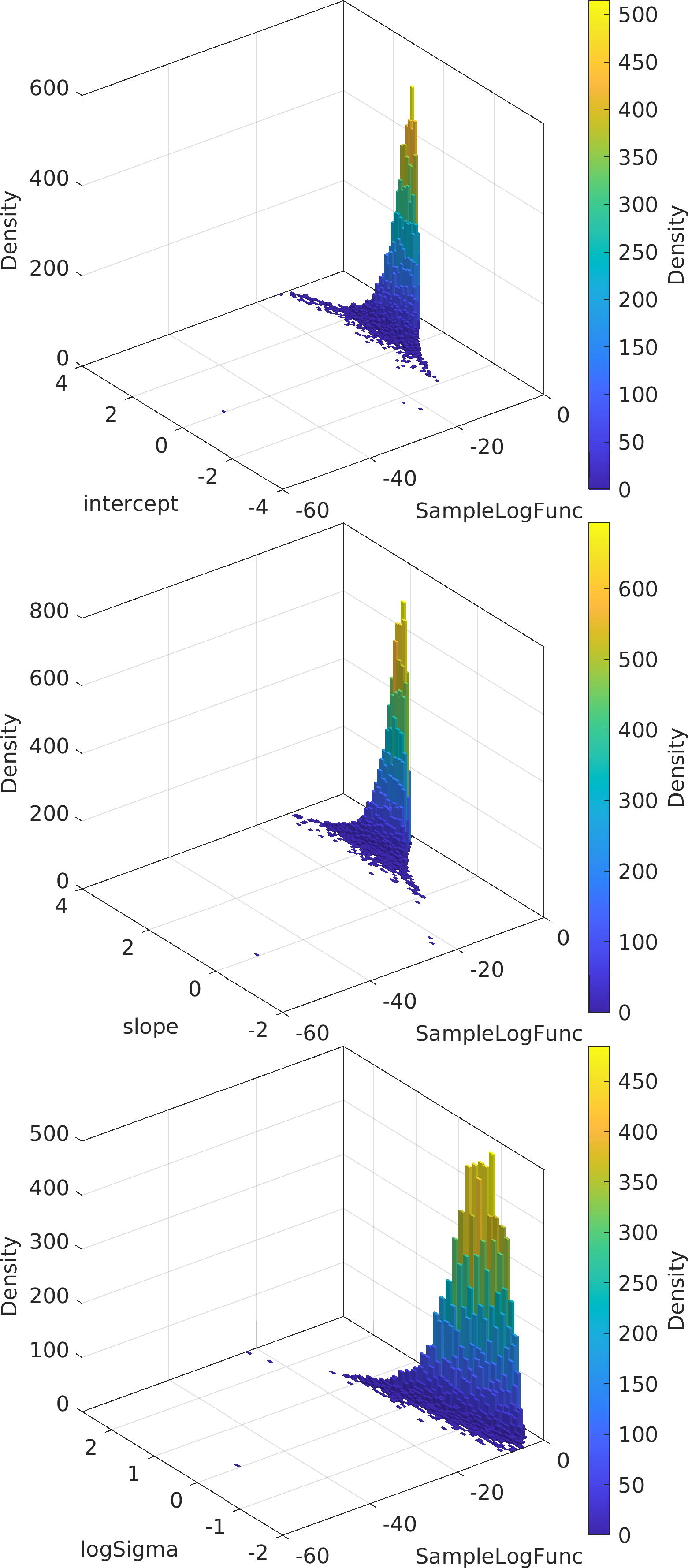
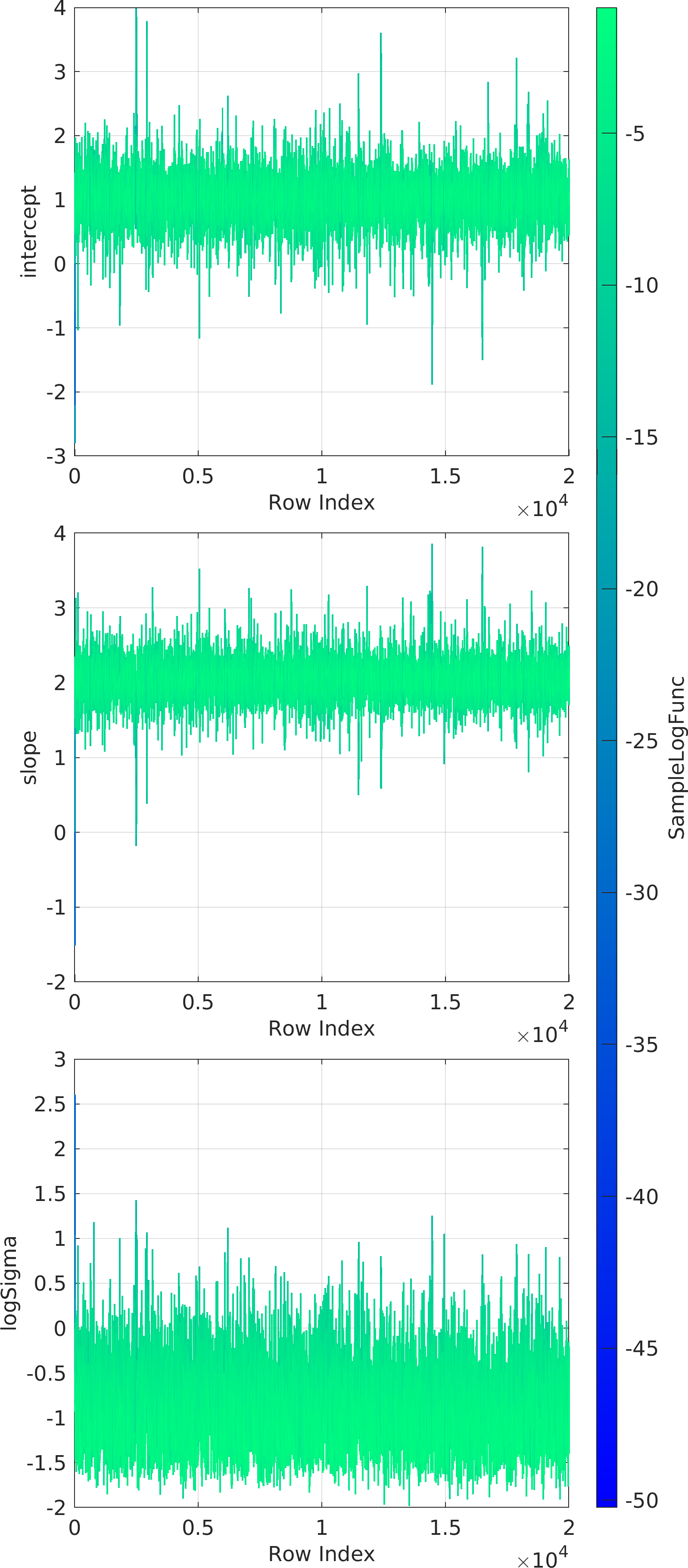
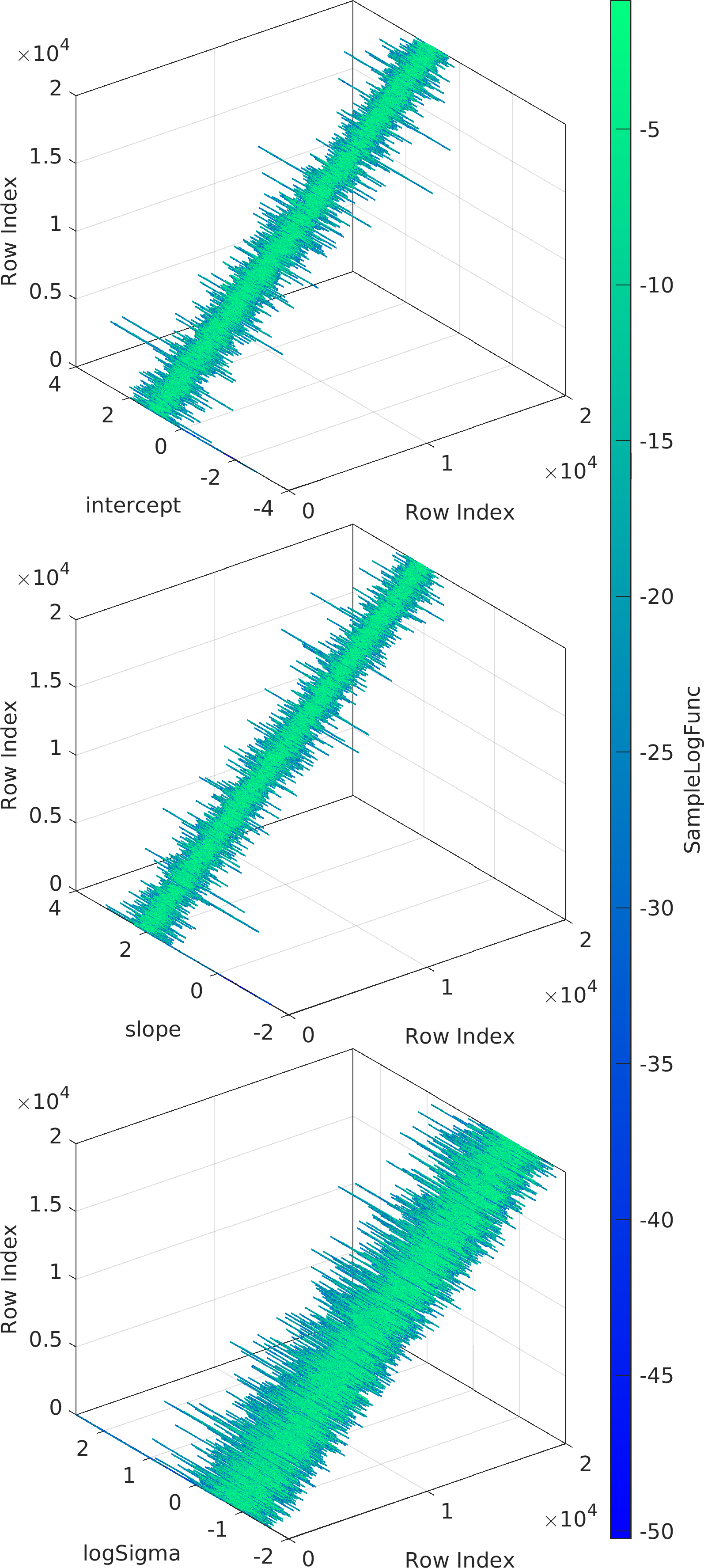
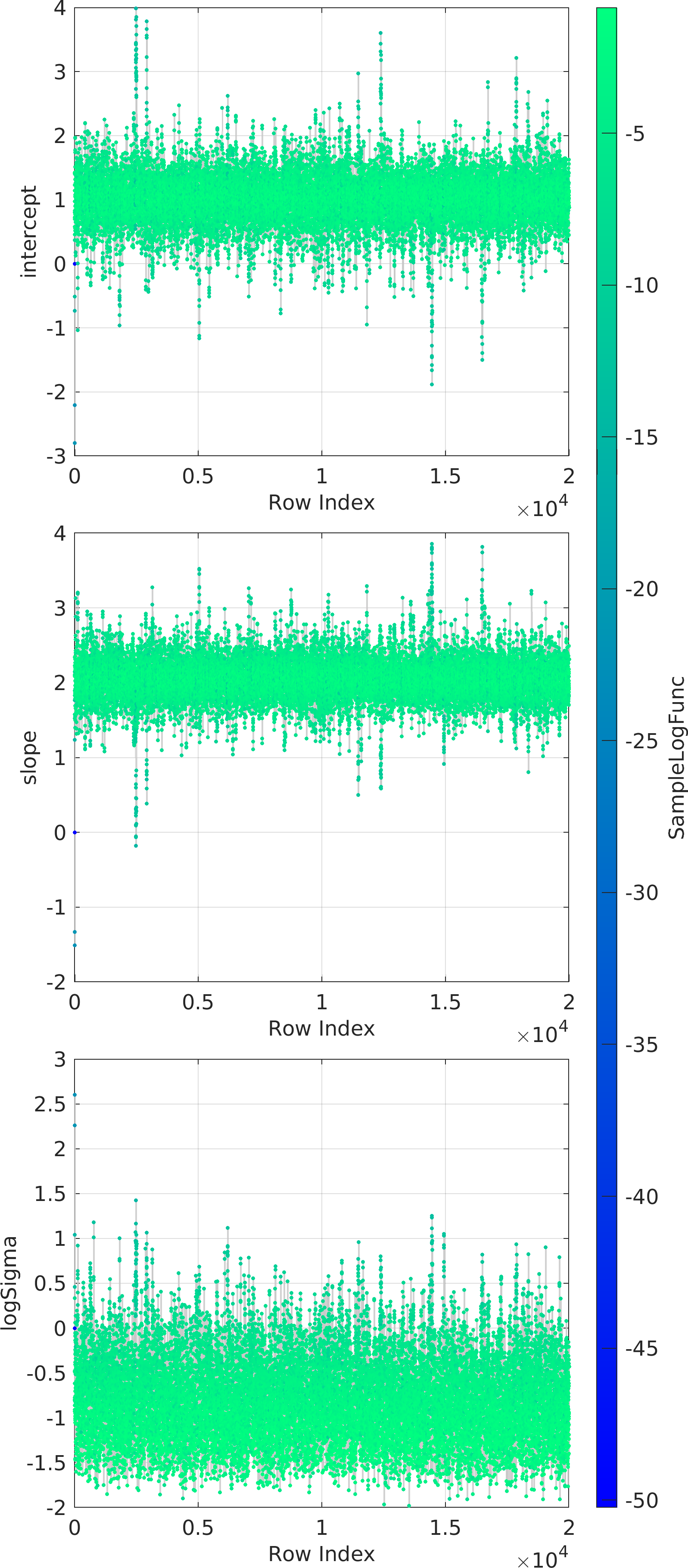
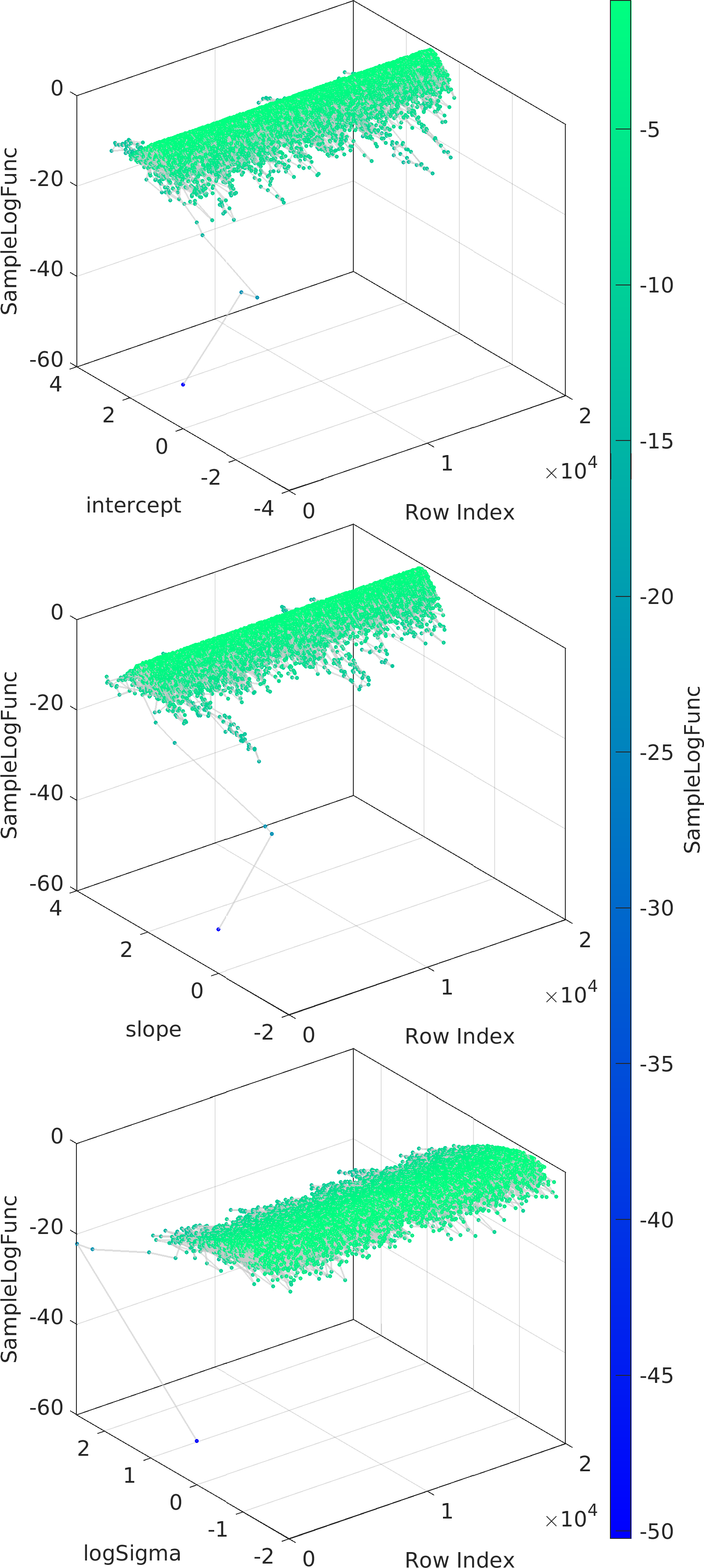
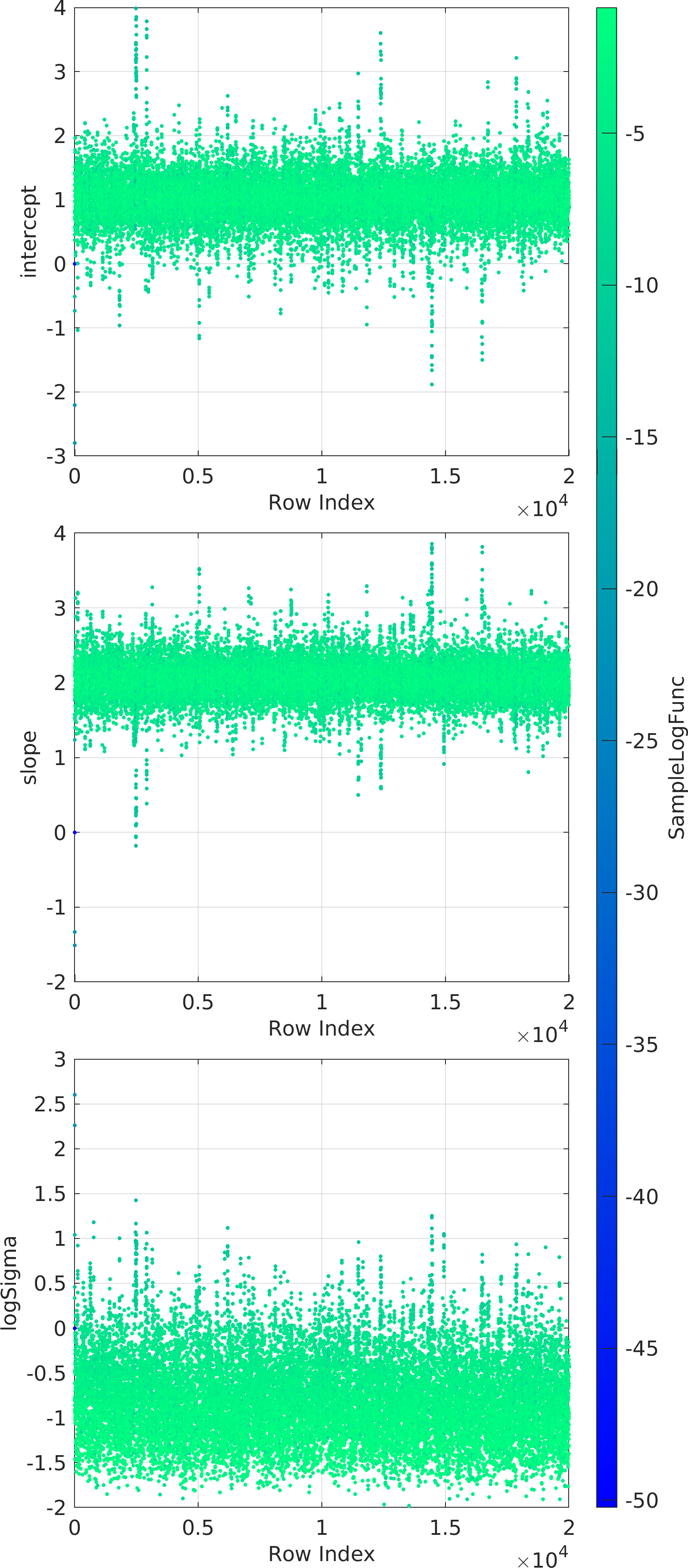
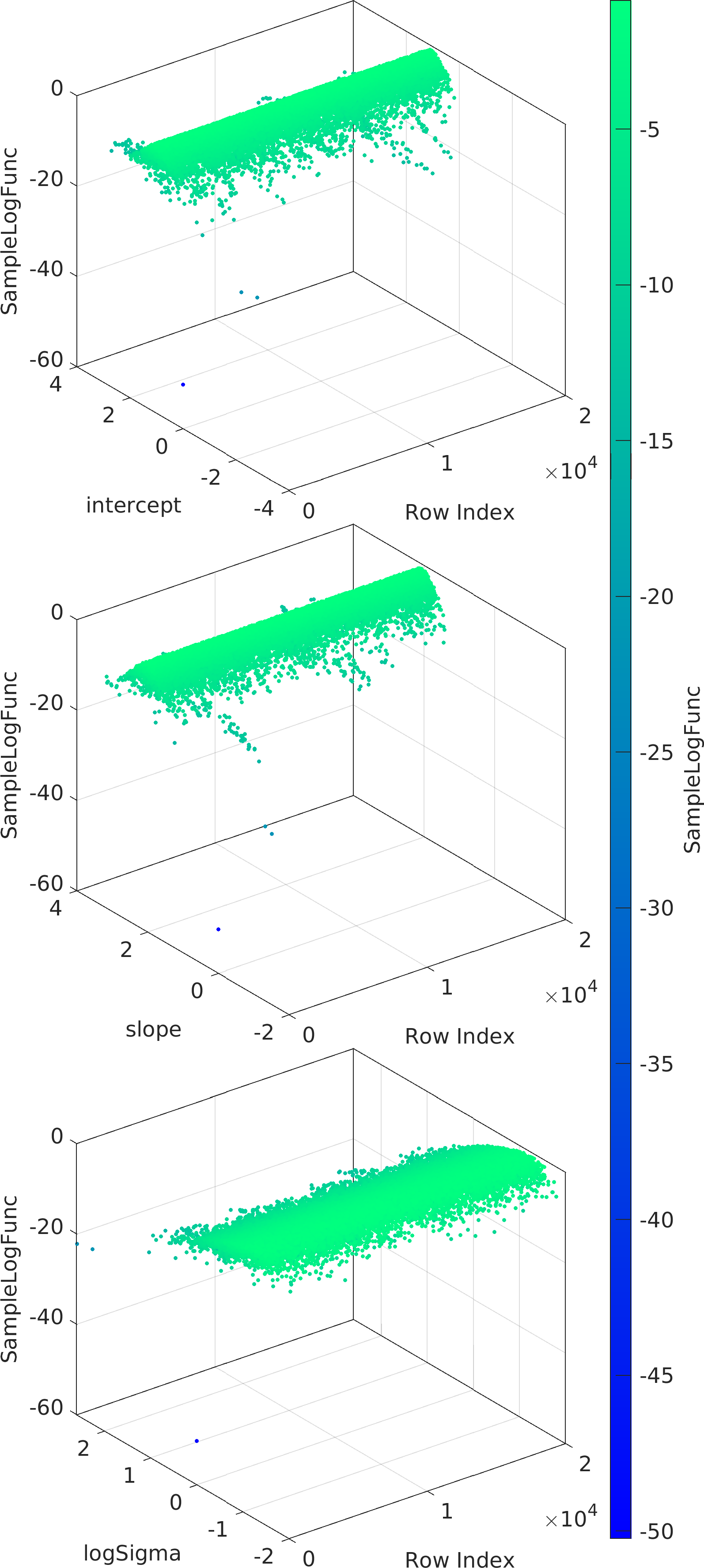
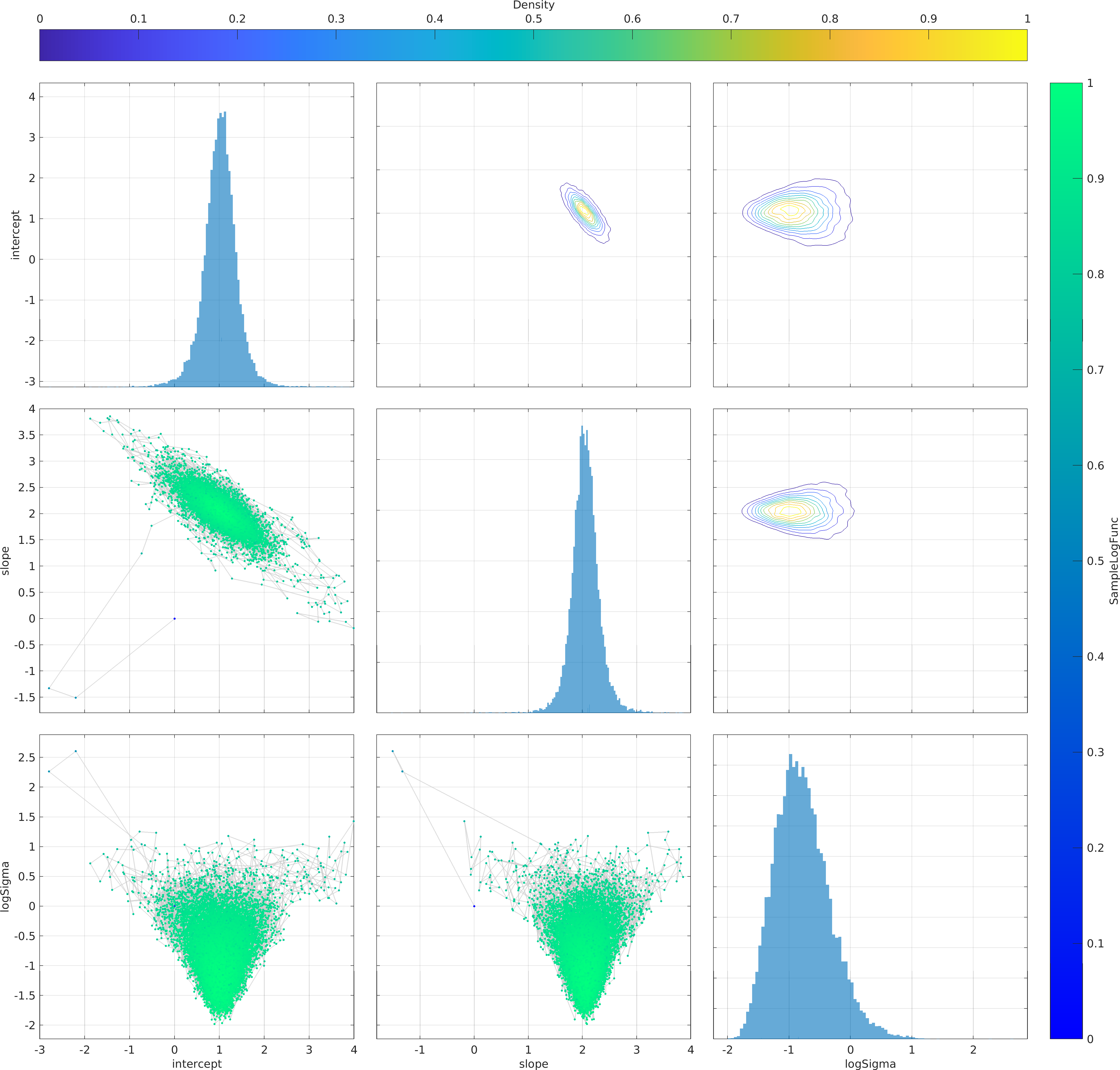
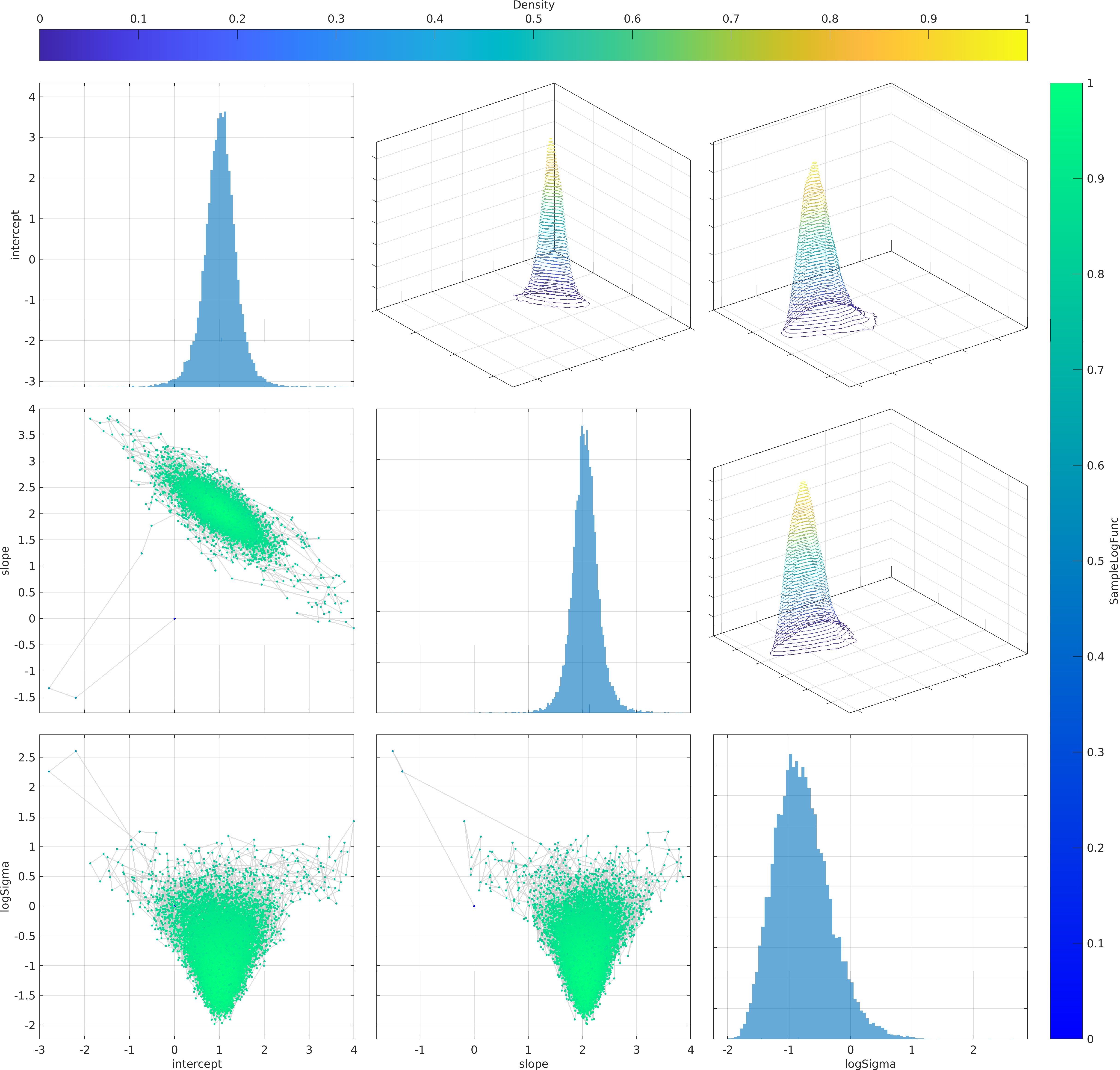
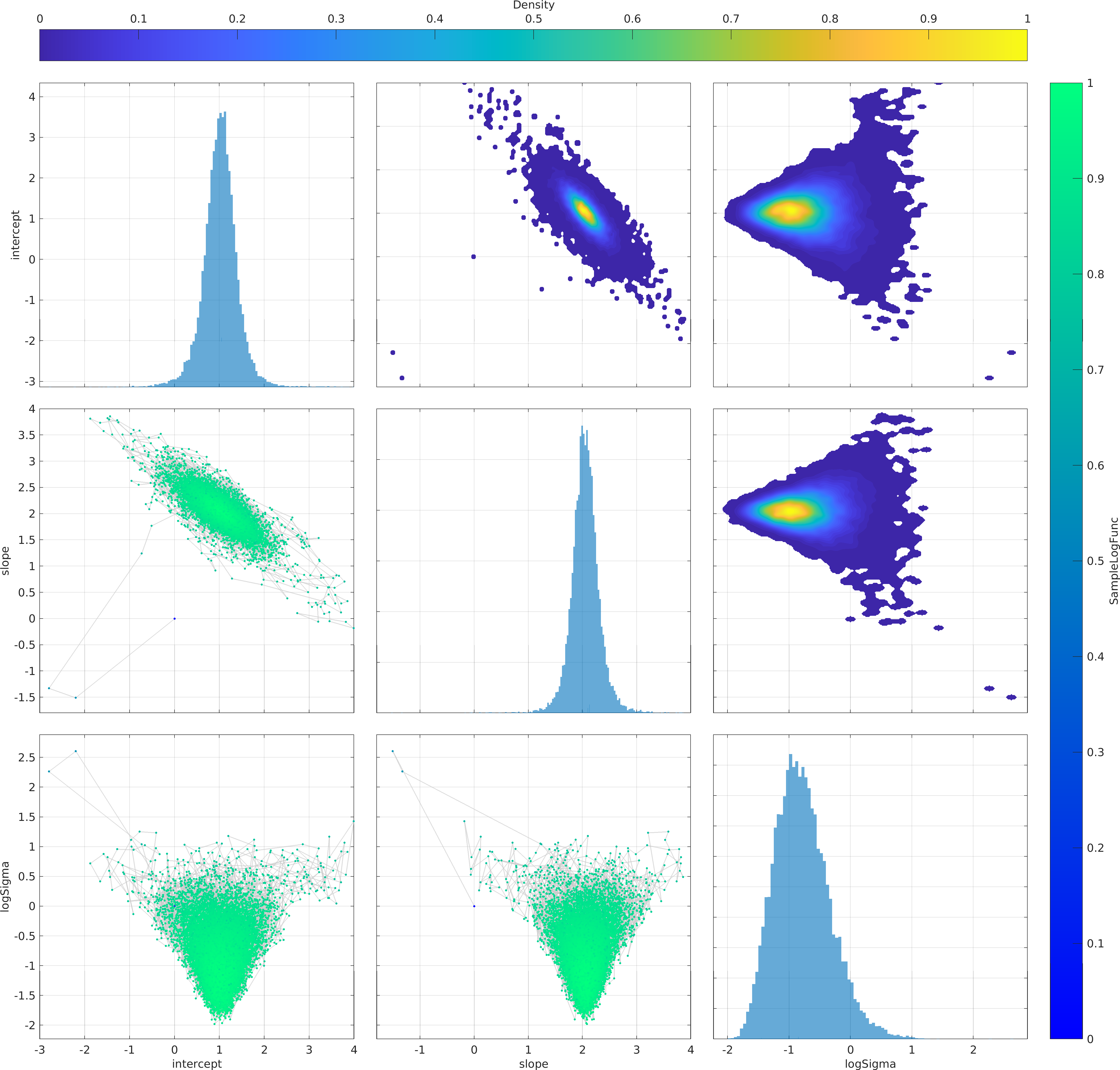
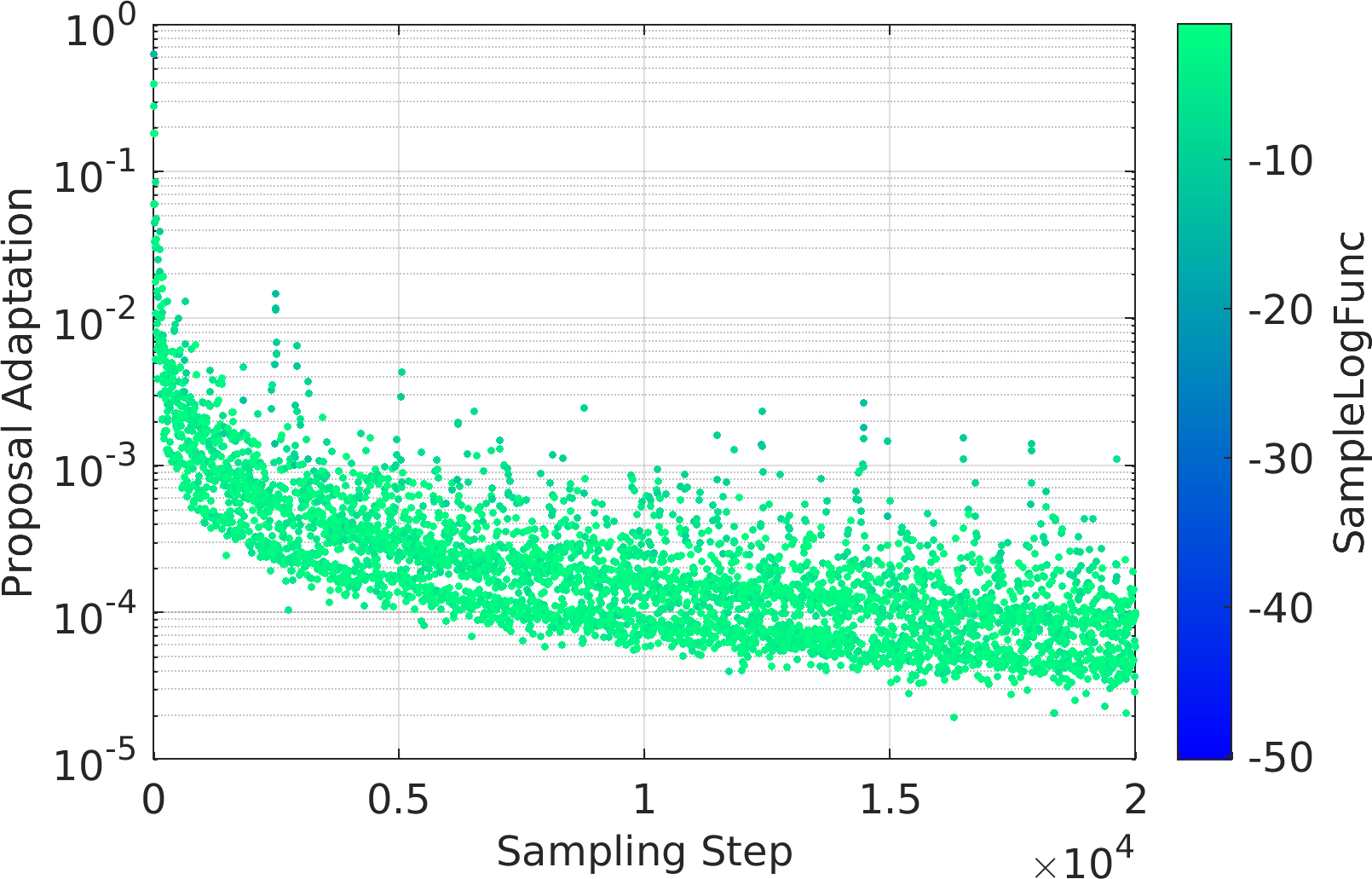
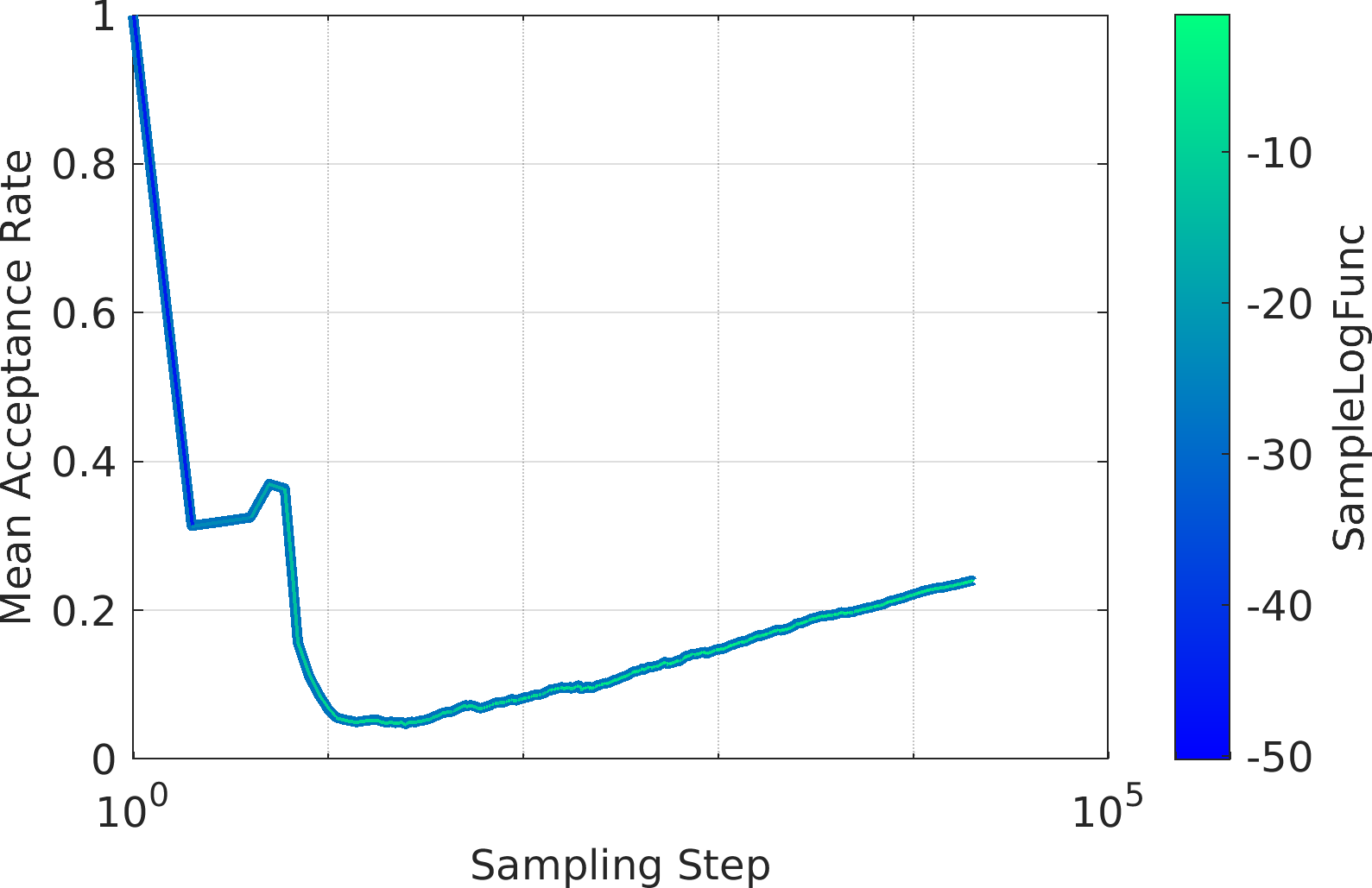
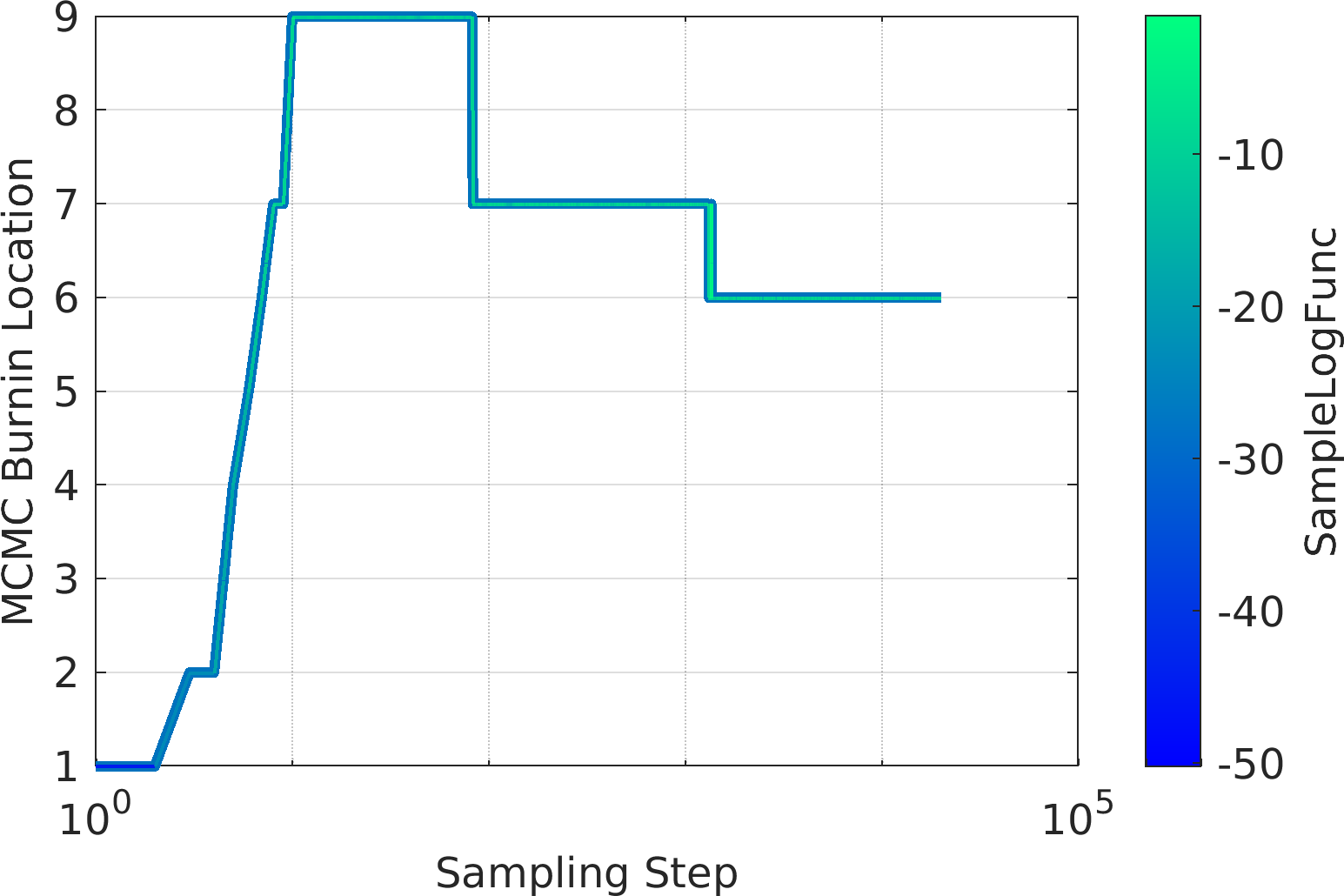
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.