|
ParaMonte MATLAB 3.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
|
ParaMonte MATLAB 3.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
This is the ParaDRAM class for generating instances of serial and parallel Delayed-Rejection Adaptive Metropolis-Hastings Markov Chain Monte Carlo sampler of the ParaMonte MATLAB library.
More...
Public Member Functions | |
| function | Paradram () |
| Generate and return an instance of the serial and parallel Delayed-Rejection Adaptive Metropolis-Hastings Markov Chain Monte Carlo sampler of the ParaMonte MATLAB library. More... | |
| function | getppm (in self) |
| Generate and return the relevant Post-Processing Message (ppm) for the current ParaMonte sampler to be displayed on the MATLAB command line after the sampling is complete. More... | |
| function | readChainMarkov (in self, in pattern, in sep) |
Return a list of objects of class pm.sampling.FileContentsChainDRAM containing the content(s) of the ParaMonte simulation output chain file(s) whose path(s) match the specified input pattern or the simulation specification sampler.spec.outputFileName.More... | |
| function | run (in self, in getLogFunc, in ndim) |
| Run the ParaDRAM sampler and return nothing. More... | |
This is the ParaDRAM class for generating instances of serial and parallel Delayed-Rejection Adaptive Metropolis-Hastings Markov Chain Monte Carlo sampler of the ParaMonte MATLAB library.
For more information, see the documentation of the class constructor pm.sampling.Paradram::Paradram.
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
Definition at line 17 of file Paradram.m.
| function Paradram::Paradram | ( | ) |
Generate and return an instance of the serial and parallel Delayed-Rejection Adaptive Metropolis-Hastings Markov Chain Monte Carlo sampler of the ParaMonte MATLAB library.
This function is the constructor of the pm.sampling.Paradram sampler class.
Once you assign the desired simulation specifications to the corresponding attributes within the component spec of an object of class pm.sampling.Paradram, call the ParaDRAM sampler via the object method pm.sampling.Paradram.run().
While the constructor of this class does not take any input arguments, all ParaDRAM simulation specifications can be set after creating the object.
sampler : The output scalar object of class pm.sampling.Paradram.
Possible calling interfaces ⛓
[] or re-instantiate an object of class pm.sampling.Paradram.run().The ParaDRAM simulation specifications have lengthy comprehensive descriptions that appear in full in the output report files of every ParaDRAM simulation.
The best way to learn about individual ParaDRAM simulation attributes is to a run a minimal serial simulation as given in the above.
You can also use the sampler.spec.doc() method:
First, ensure the ParaMonte +pm package (i.e., folder) is available in your MATLAB paths.
Here is a MATLAB script main.m for a serial ParaDRAM simulation.
Copy and paste the following code into your MATLAB session:
The mathematical objective function in the above example is a is a multivariate Normal distribution centered at the origin, whose natural logarithm is returned by the lambda (Anonymous) function defined as a function handle input to the ParaDRAM sampler.
Running this code will generate a set of simulation output files (in the current working directory of MATLAB).
Among these, the file suffixed with "_report.txt" contains the full description of all input specifications of the ParaDRAM simulation as well as other information about the simulation results.
First, ensure the ParaMonte +pm package (i.e., folder) is available in your MATLAB paths.
Threading parallelism is possible as of ParaMonte MATLAB version 3.0.0.
However, only singleChain ParaDRAM simulations are supported.
Here is a MATLAB script main.m for a thread-parallel ParaDRAM simulation.
Copy and paste the following code and paste into your MATLAB session:
The statement sample.vis.tile.line.make(); in the above is equivalent to the following set of lines:
The mathematical objective function in the above example is a is a multivariate Normal distribution centered at the origin, whose natural logarithm is returned by the lambda (Anonymous) function defined as a function handle input to the ParaDRAM sampler.
Running this code will generate a set of simulation output files (in the current working directory of MATLAB).
Among these, the file suffixed with "_report.txt" contains the full description of all input specifications of the ParaDRAM simulation as well as other information about the simulation results.
Specifying 0 as the number of threads will lead to using all available CPU threads for thread-parallel ParaDRAM simulation.
First, ensure the ParaMonte +pm package (i.e., folder) is available in your MATLAB paths.
MPI-parallel simulations can be slightly more cumbersome than thread-parallel simulations described above because MPI-parallel simulations cannot be performed from within a MATLAB GUI session and require launching MATLAB via a compatible mpiexec launcher.
Ensure you need and will get a speedup by running the an MPI-parallel simulation.
Typically, your simulation may then benefit from parallelism only if a single evaluation of the objective function takes longer than a few milliseconds.
Ensure the required MPI libraries are installed on your system (You can skip this step if you know that you already have a compatible MPI library installed on your system).
On the MATLAB command line type the following,
This will verify the existence of a valid MPI library on your system and, if missing, will guide you to install the MPI library on your system.
Once the MPI installation is verified, copy and paste the following code into your MATLAB session:
main_mpi.m MATLAB script file in the current working directory of your MATLAB session.main_mpi.m) in parallel.On Windows:
From within command prompt that recognizes both MATLAB and mpiexec, ideally, the Intel dedicated command-prompt that is shipped with Intel MPI library, type the following,
-localonly.On macOS/Linux:
From within a Bash terminal that recognizes both MATLAB and mpiexec, type the following,
main_mpi.m will run on 3 processors.-batch mode for parallel simulations, disabling all plotting capabilities.
Example usage ⛓
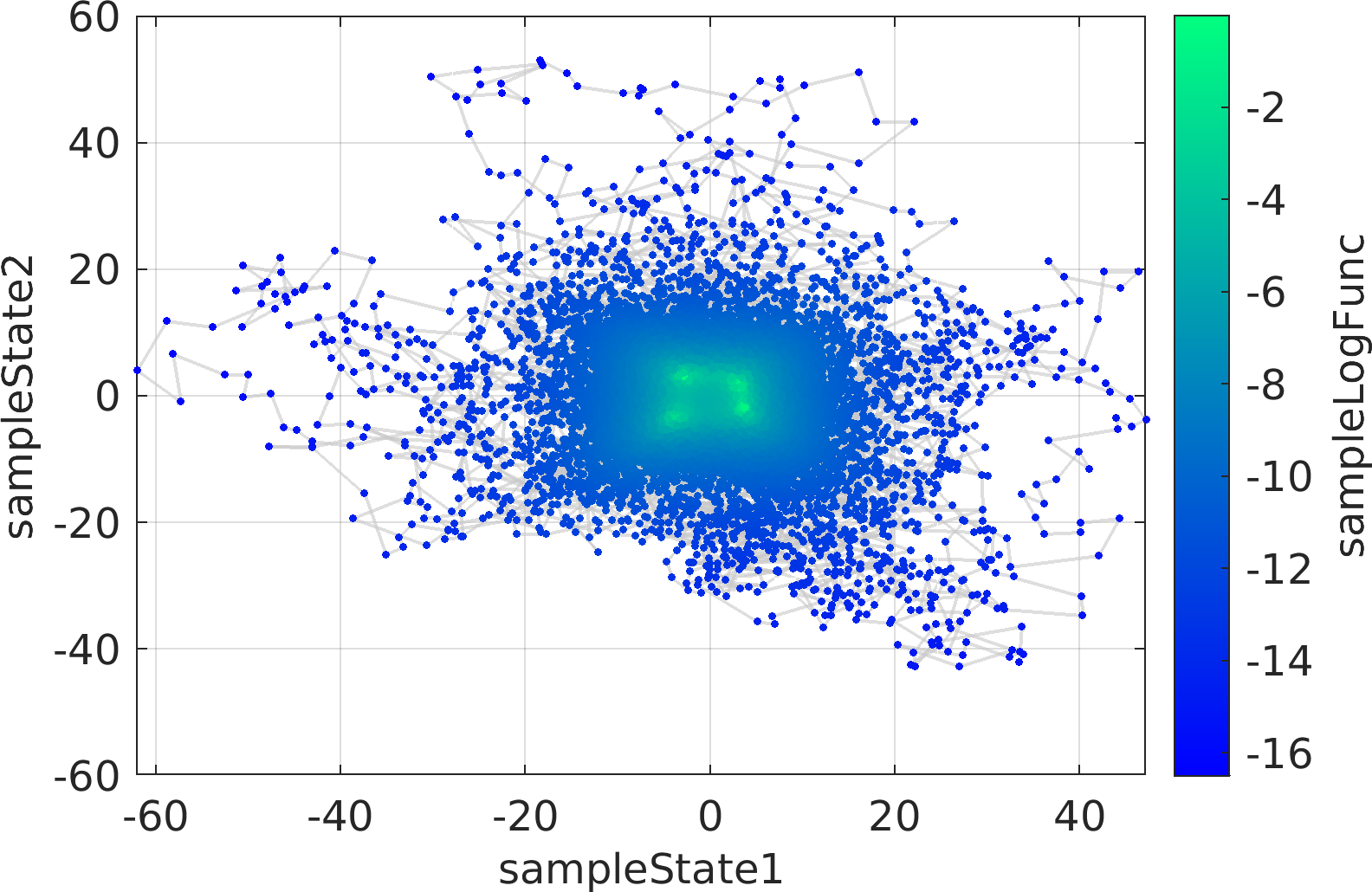
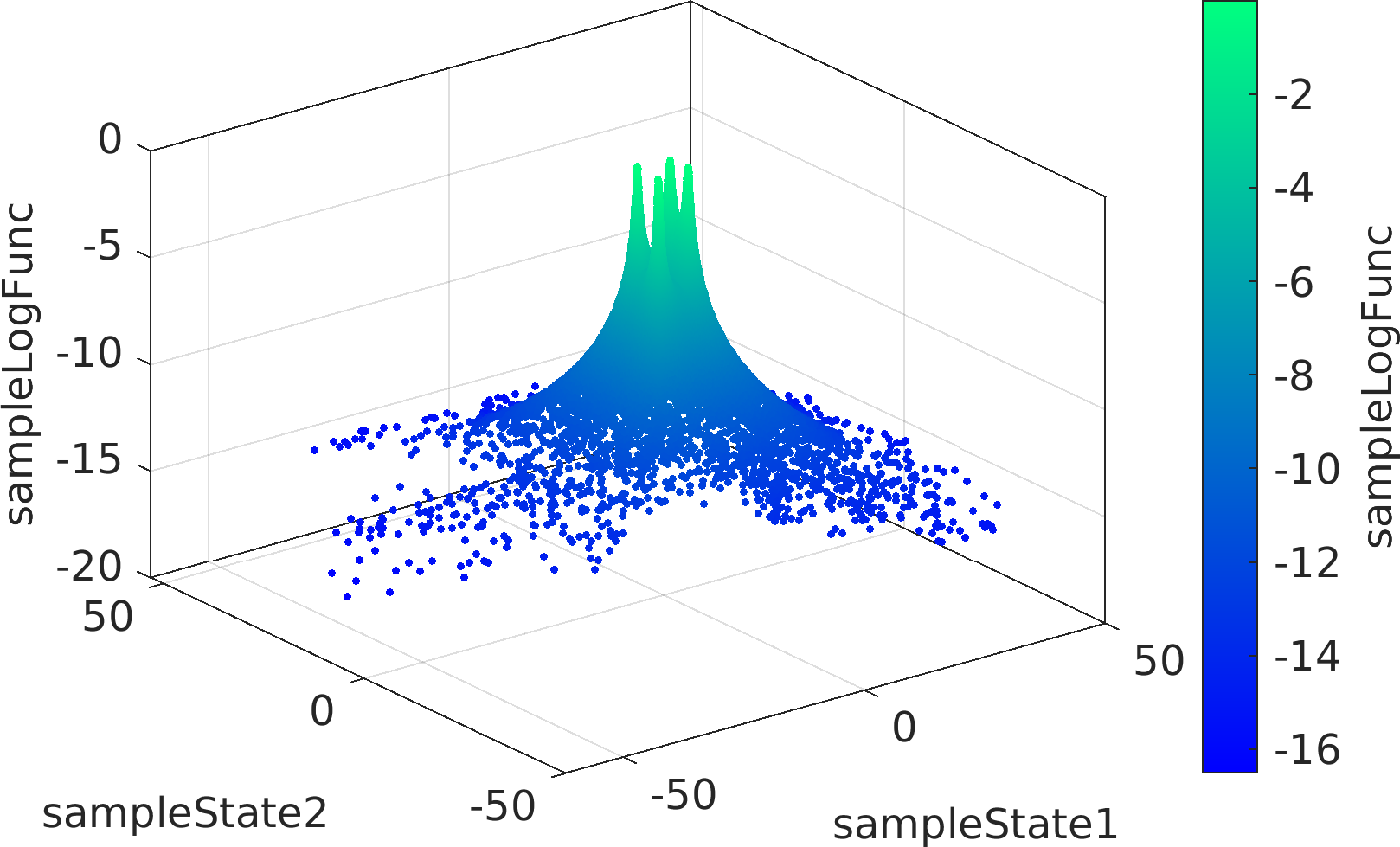
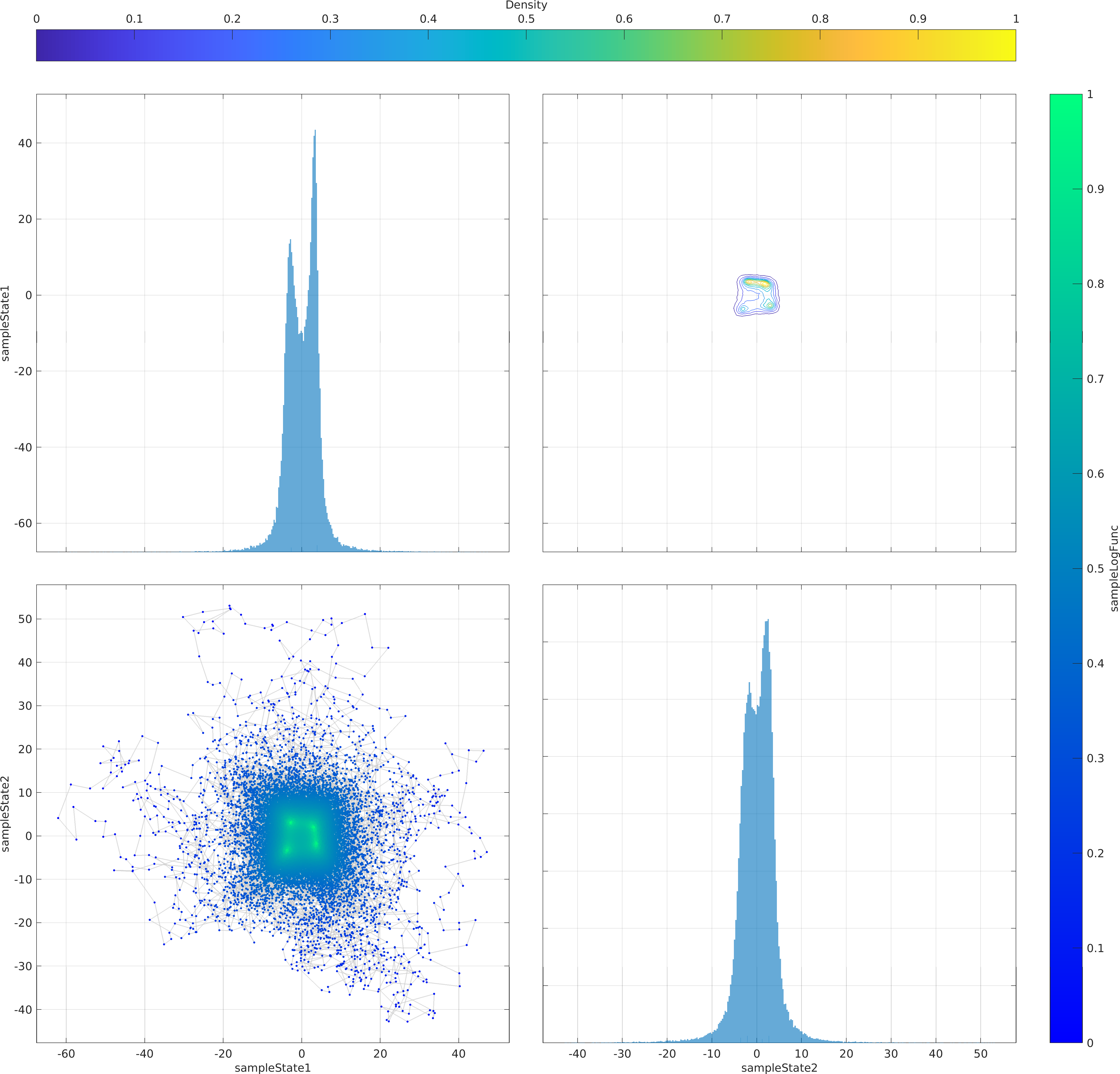
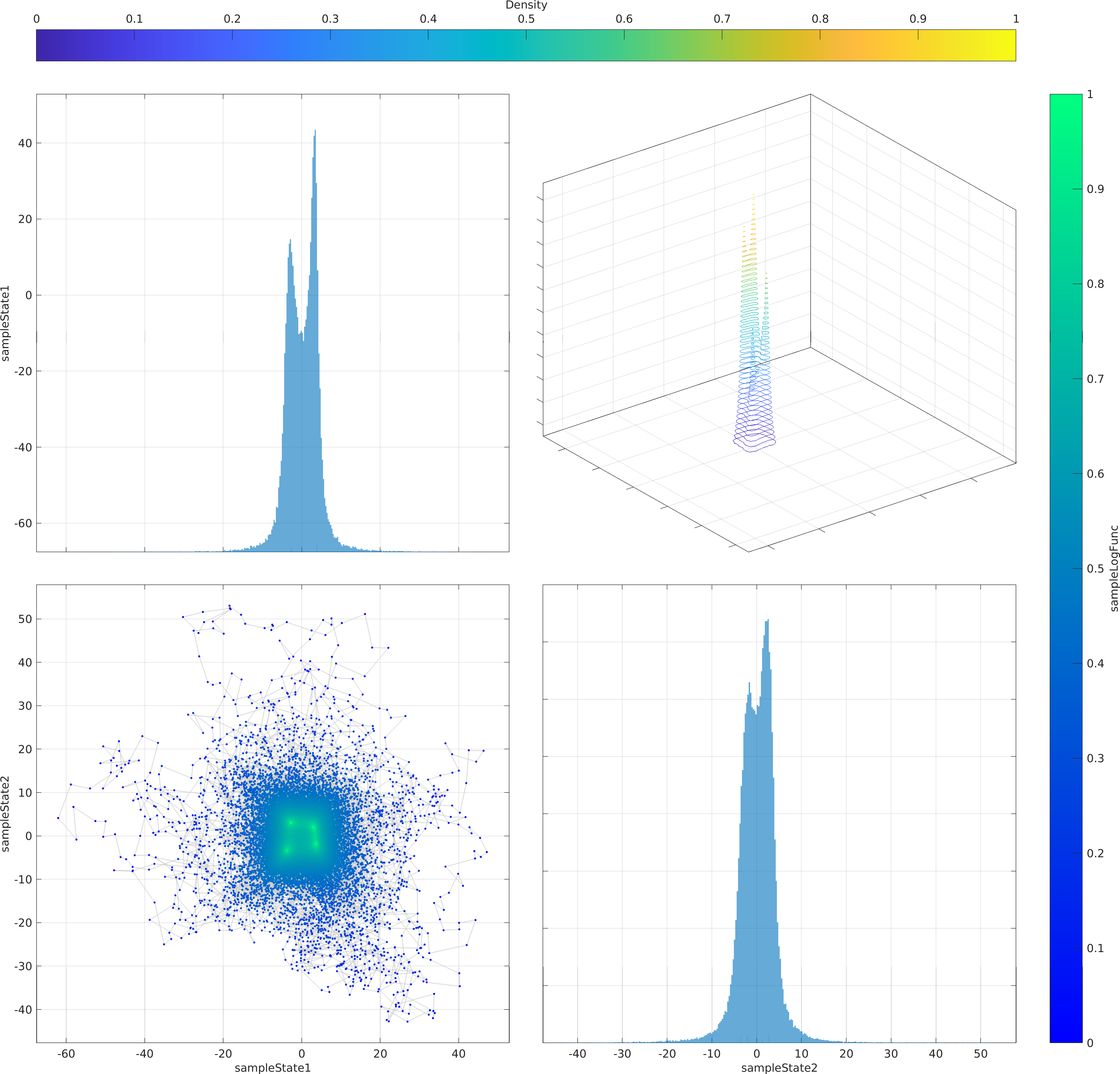
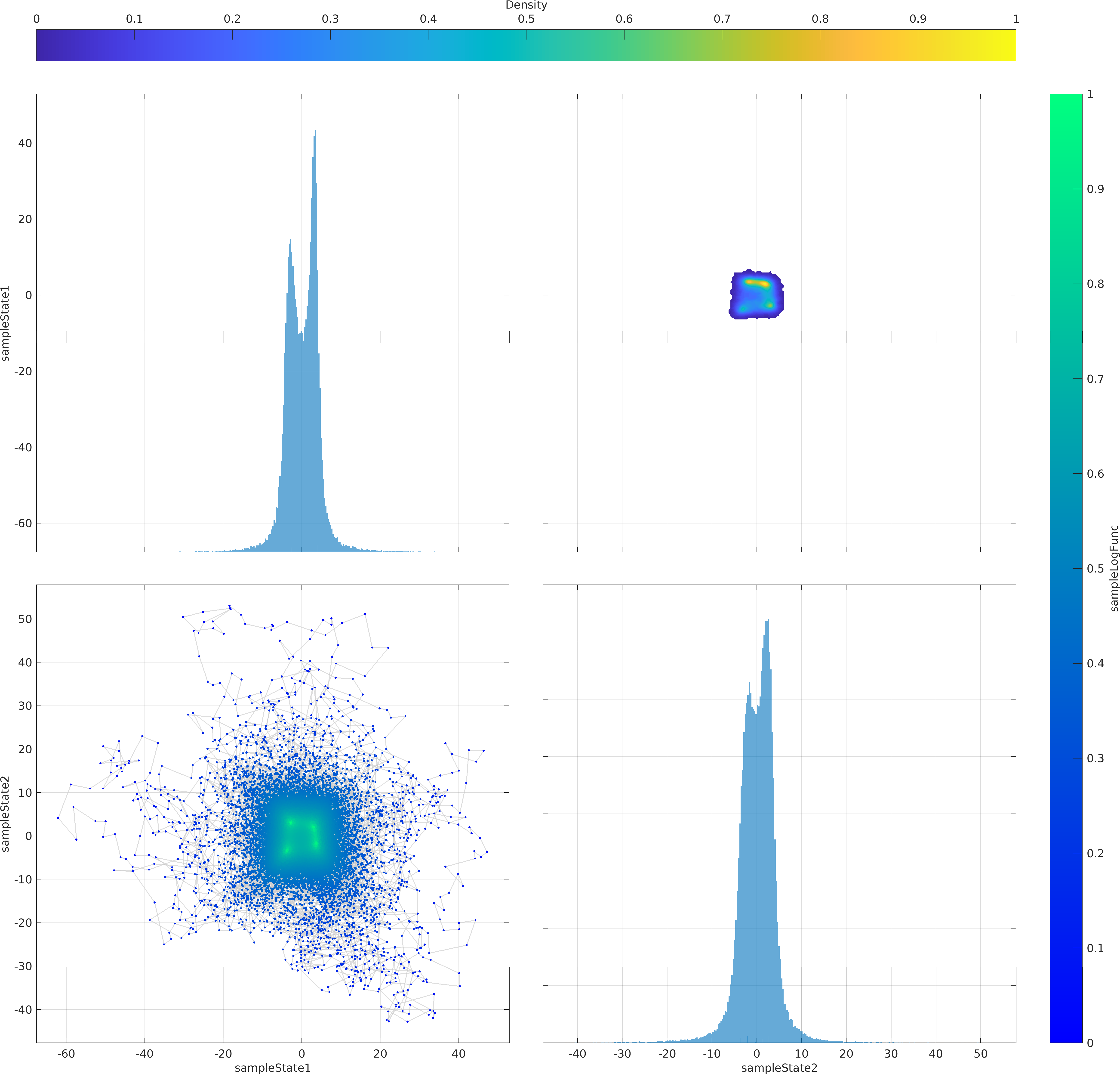
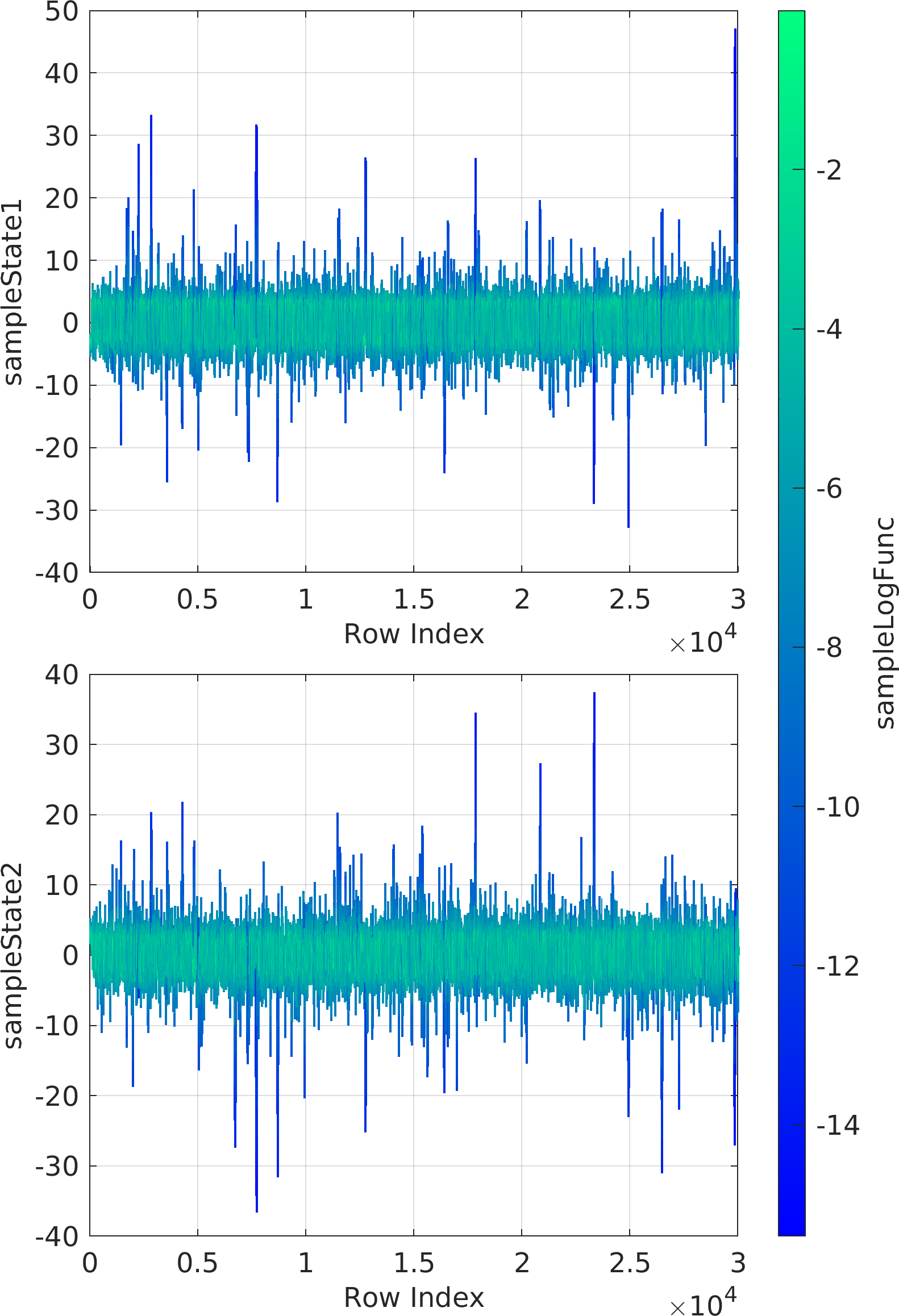
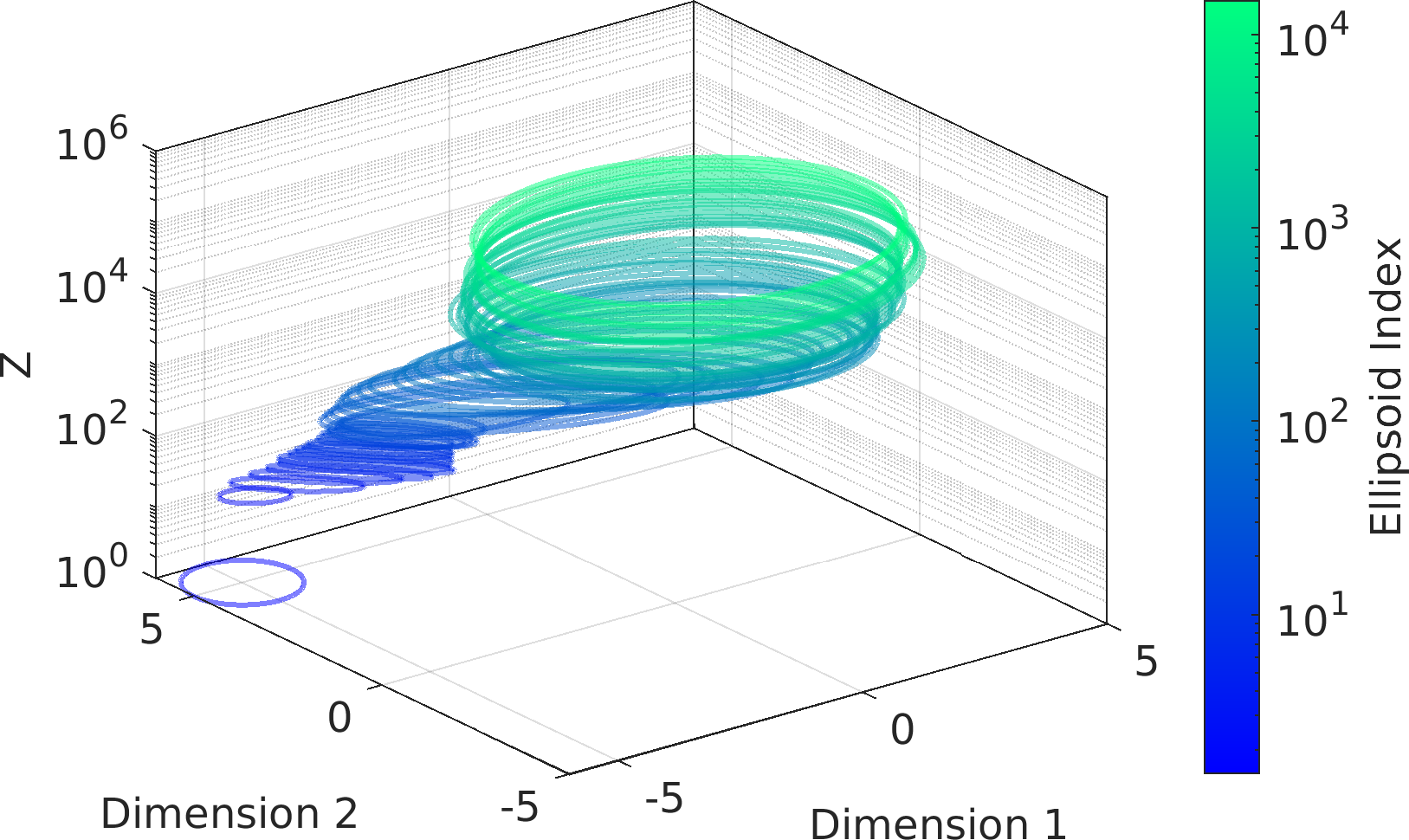
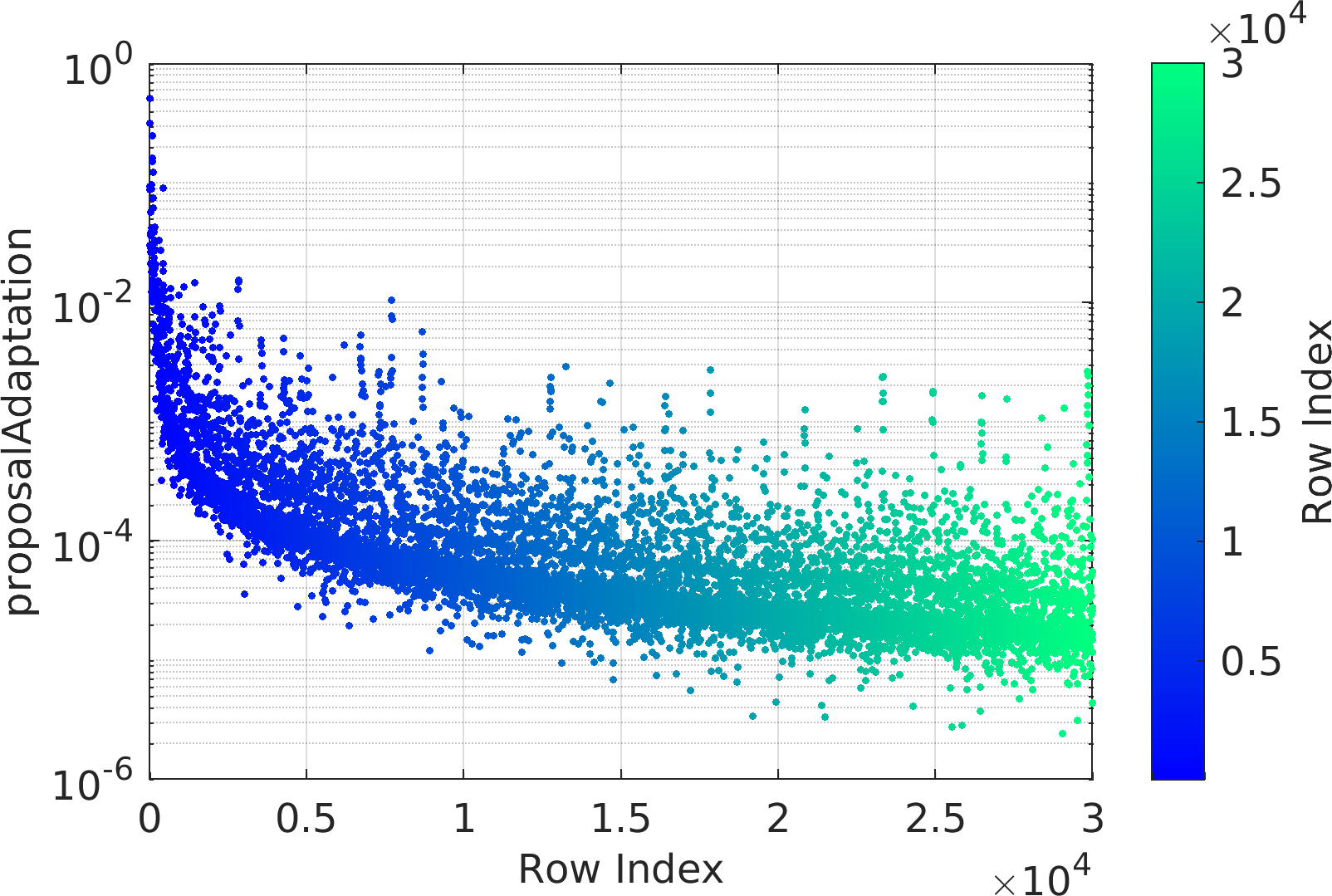
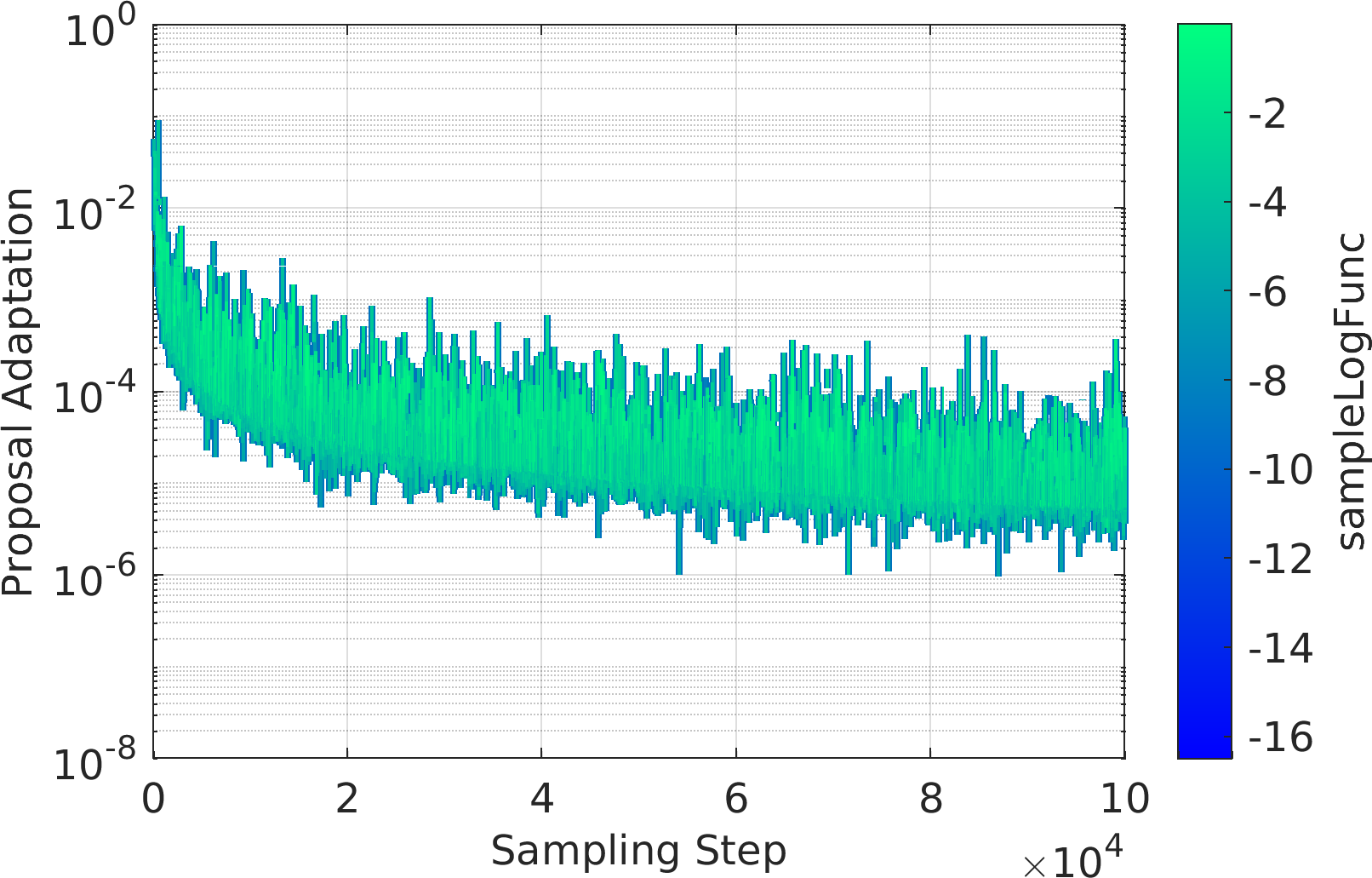
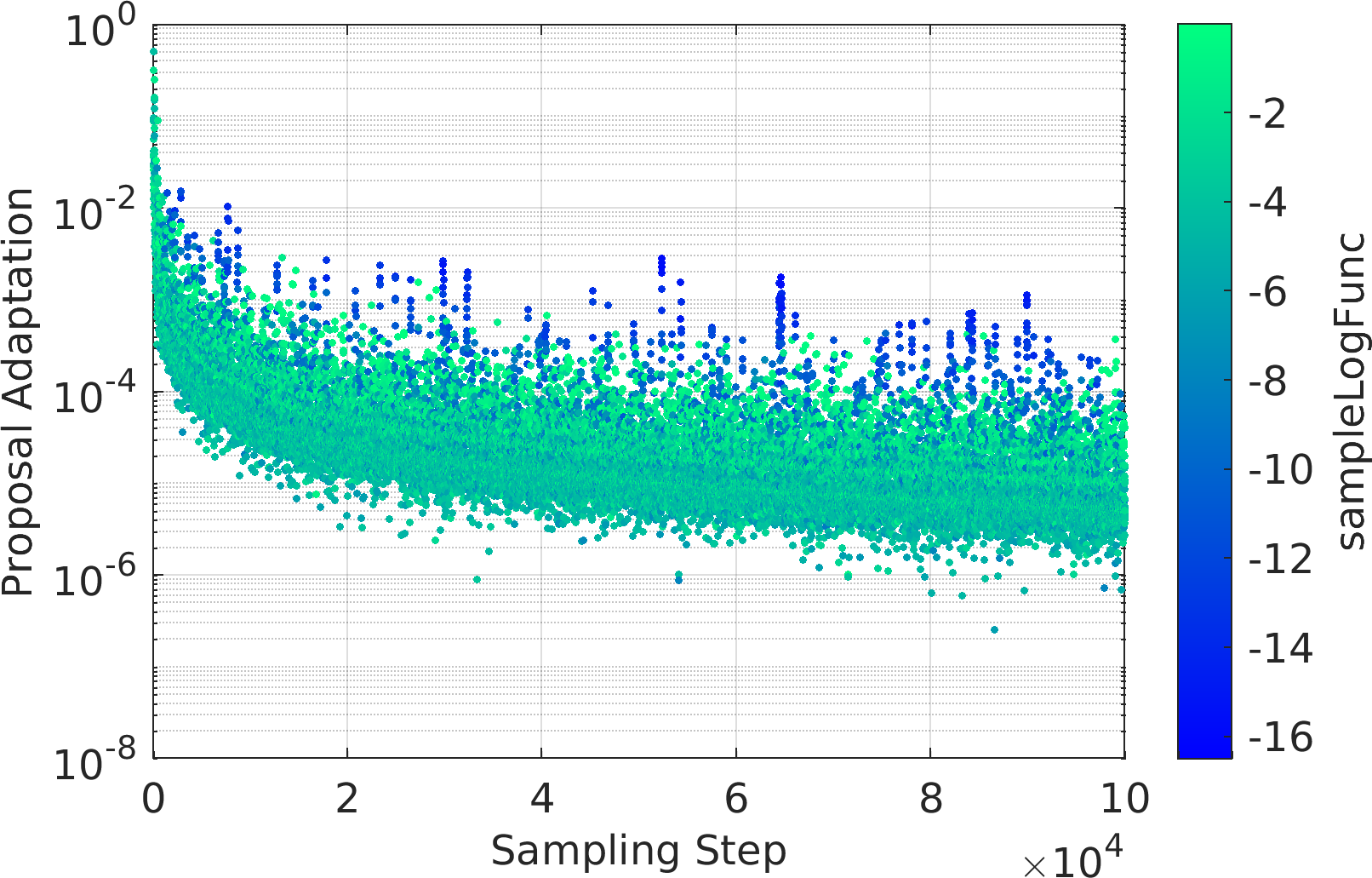
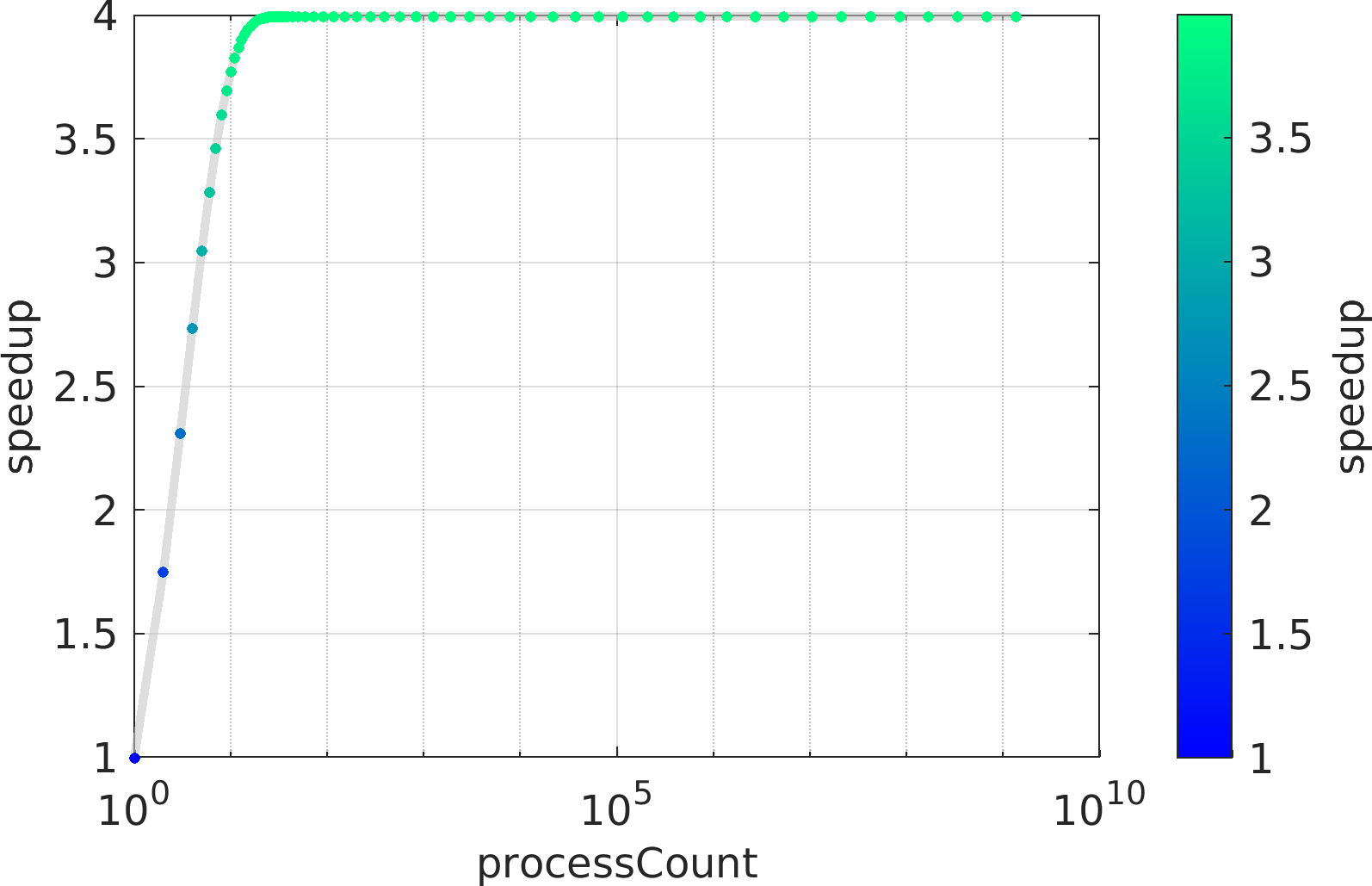
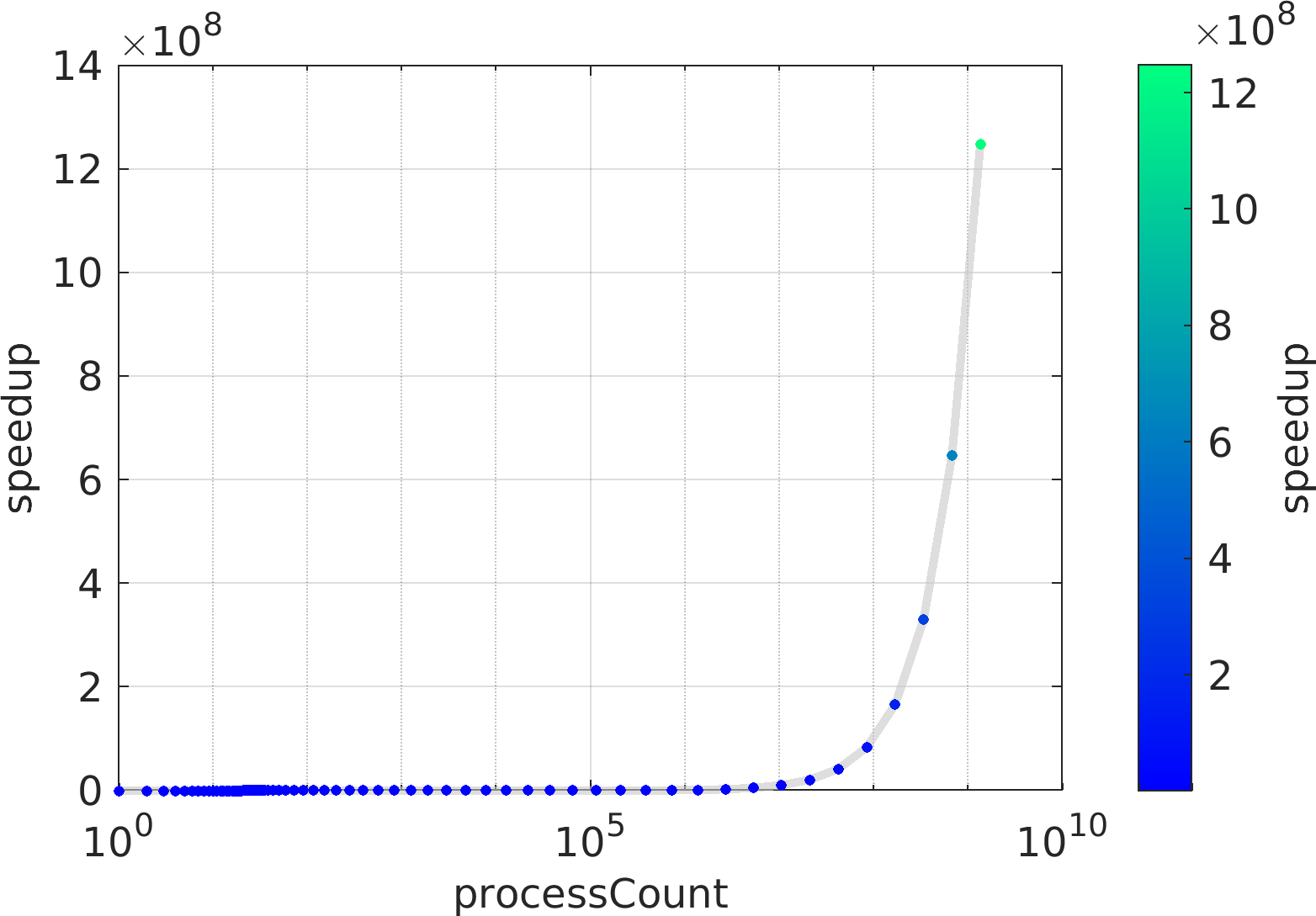
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| function Paradram::getppm | ( | in | self | ) |
Generate and return the relevant Post-Processing Message (ppm) for the current ParaMonte sampler to be displayed on the MATLAB command line after the sampling is complete.
This is a private dynamic method of the pm.sampling.Paradram sampler class.
This method is not meant to be used or accessed by the end users.
| [in] | self | : The input parent object of class pm.sampling.Sampler which is implicitly passed to this dynamic method (not by the user). |
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| function Paradram::readChainMarkov | ( | in | self, |
| in | pattern, | ||
| in | sep | ||
| ) |
Return a list of objects of class pm.sampling.FileContentsChainDRAM containing the content(s) of the ParaMonte simulation output chain file(s) whose path(s) match the specified input pattern or the simulation specification sampler.spec.outputFileName.
| [in] | self | : The input/output parent object of class pm.sampling.Paradram which is implicitly passed to this dynamic method (not by the user). |
| [in] | pattern | : See the documentation of the corresponding argument of the constructor of the method pm.sampling.Sampler.readChain. (optional, The default is set by pm.sampling.Sampler.readChain) |
| [in] | sep | : See the documentation of the corresponding argument of the constructor of the method pm.sampling.Sampler.readChain. (optional, The default is set by pm.sampling.Sampler.readChain) |
chainMarkovList : The output cell array of objects of class pm.sampling.FileContentsChainDRAM, each of which corresponds to the contents of a unique chain file.doc pm.sampling.FileContentsChainDRAM to see the documentation of the contents of the objects of the output list.
Possible calling interfaces ⛓
Example usage ⛓
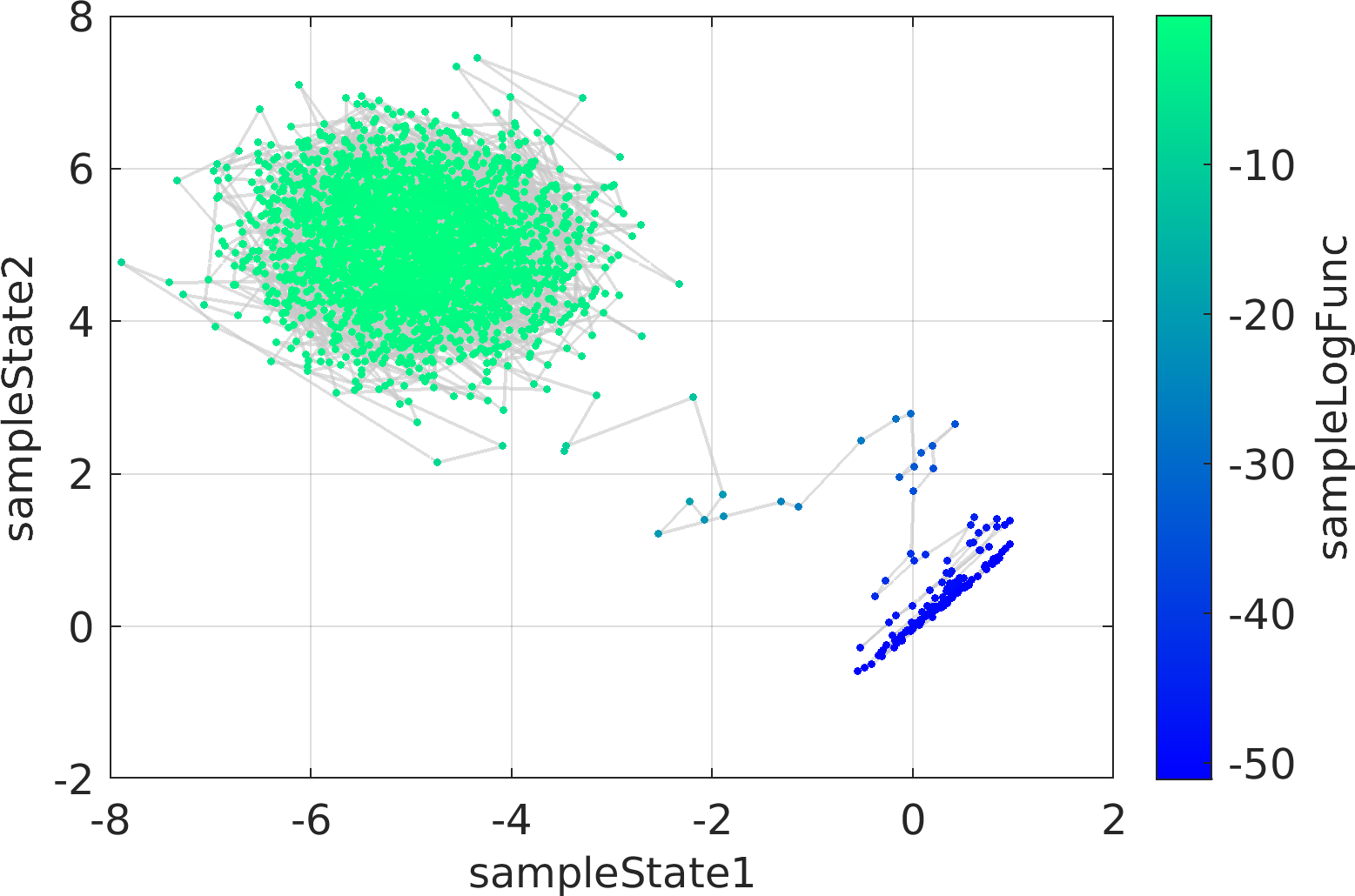
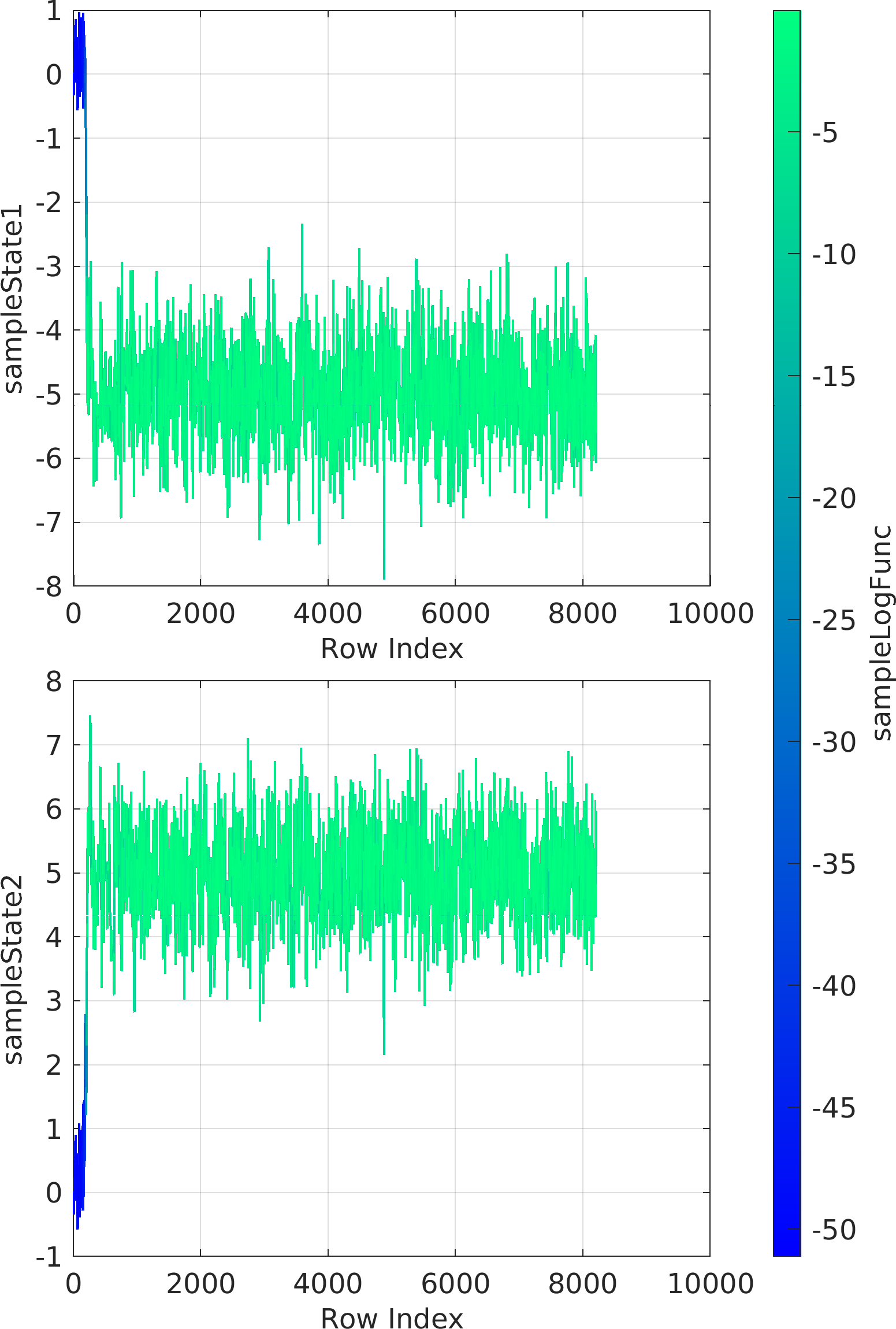
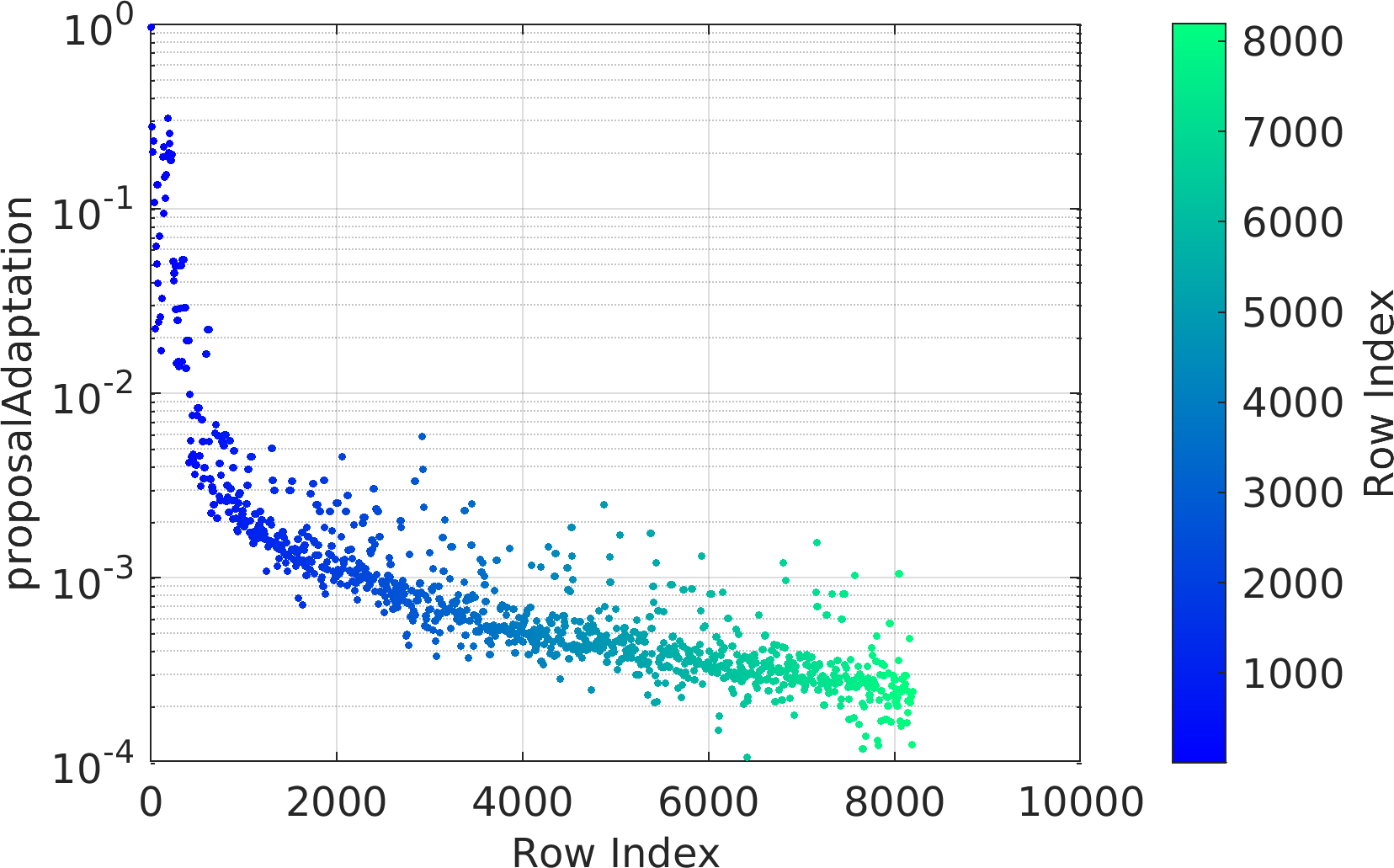
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
| function Paradram::run | ( | in | self, |
| in | getLogFunc, | ||
| in | ndim | ||
| ) |
Run the ParaDRAM sampler and return nothing.
For example usage, see the documentation of the parent class of this method pm.sampler.Paradram.
| [in,out] | self | : The input/output parent object of class pm.sampling.Paradram which is implicitly passed to this dynamic method (not by the user). |
| [in] | getLogFunc | : The input MATLAB function handle or anonymous (lambda) function containing the implementation of the objective function to be sampled. This user-specified function must have the following interface, function logFunc = getLogFunc(state)
end
|
| [in] | ndim | : The input scalar positive-valued whole-number representing the number of dimensions of the domain of the user-specified objective function in the input getLogFunc(). |
Possible calling interfaces ⛓
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.