
|
ParaMonte Fortran 2.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |

|
ParaMonte Fortran 2.0.0
Parallel Monte Carlo and Machine Learning Library
See the latest version documentation. |
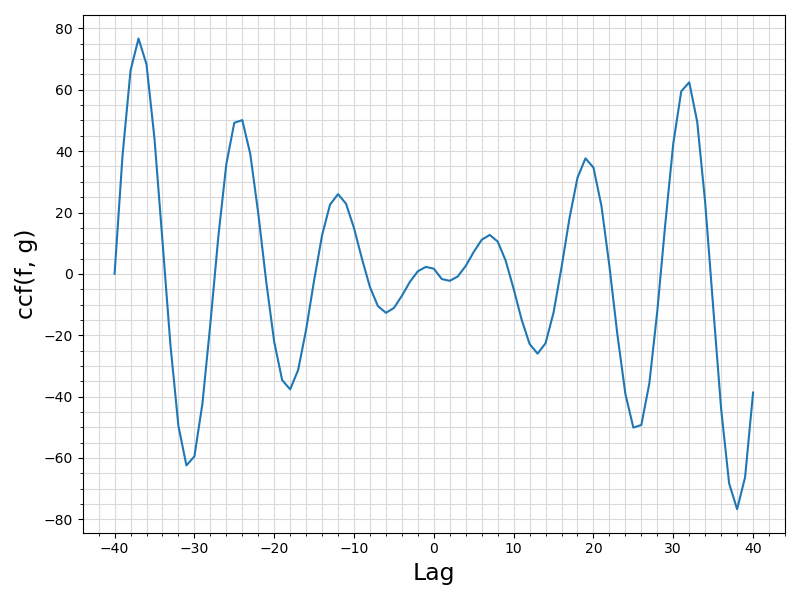
Return the cross-correlation function (CCF) \((f\star g)(\tau)\) of the discrete signal \(g\) lagging the signal the discrete signal \(f\) for a range of lags that spans the maximum of the lengths of the two sequences.
More...
Return the cross-correlation function (CCF) \((f\star g)(\tau)\) of the discrete signal \(g\) lagging the signal the discrete signal \(f\) for a range of lags that spans the maximum of the lengths of the two sequences.
See the documentation of the parent module pm_sampleCCF for algorithmic details and sample correlation matrix definition.
size(f) + size(g) - 1.1 / size(ccf) / sqrt(sum(abs(f)**2) + sum(abs(g)**2)) to properly normalize the output CCF to the range [-1, +1].1 / size(ccf).size(f) + size(g) - 1, then, ccf(1 : size(g)) by this generic interface contains the cross-correlation corresponding to lags [(i, i = 0, size(g))]. ccf(size(g) + 1 : size(ccf)) contains the cross-correlation corresponding to lags [(i, i = -size(f) + 1, -1)]. | [in] | factor | : The input contiguous vector of shape (:) of type integer of default kind IK, containing the factorization of the length of the input sequence f whose FFT is to be computed.This input argument along with the input argument coef is the direct output of getFactorFFT. |
| [in] | coef | : The input contiguous vector of shape (1:size(data)) of the same type and kind as the input argument f, containing the trigonometric look up table required for FFT of the specified sequences.This input argument along with factor is the direct output of getFactorFFT. |
| [in,out] | f | : The input contiguous vector of arbitrary size (of minimum 2) of,
On output, the contents of f are destroyed.If the output inf = .true., then f contains the resulting unnormalized CCF of the input sequences. |
| [in,out] | g | : The input contiguous vector of the same type and kind and size as the input argument f, containing the second sequence in the cross-correlation computation.On output, the contents of g are destroyed.If the condition inf = .false. holds on output, then g contains the resulting unnormalized CCF of the input sequences. |
| [out] | work | : The output contiguous vector of the same type, kind, and size as the input f, used as a workspace. |
| [out] | inf | : The output scalar of type logical of default kind LK.
|
Possible calling interfaces ⛓
2 < size(f) must hold for the corresponding input arguments.size(f) == size(g) must hold for the corresponding input arguments.size(f) == size(coef) must hold for the corresponding input arguments.size(f) == size(work) must hold for the corresponding input arguments.CHECK_ENABLED=1.pure procedure(s) documented herein become impure when the ParaMonte library is compiled with preprocessor macro CHECK_ENABLED=1.pure in release build and impure in debug and testing builds.
Example usage ⛓
ifort compiler ⛓ ifort compiler ⛓ gfortran compiler ⛓ 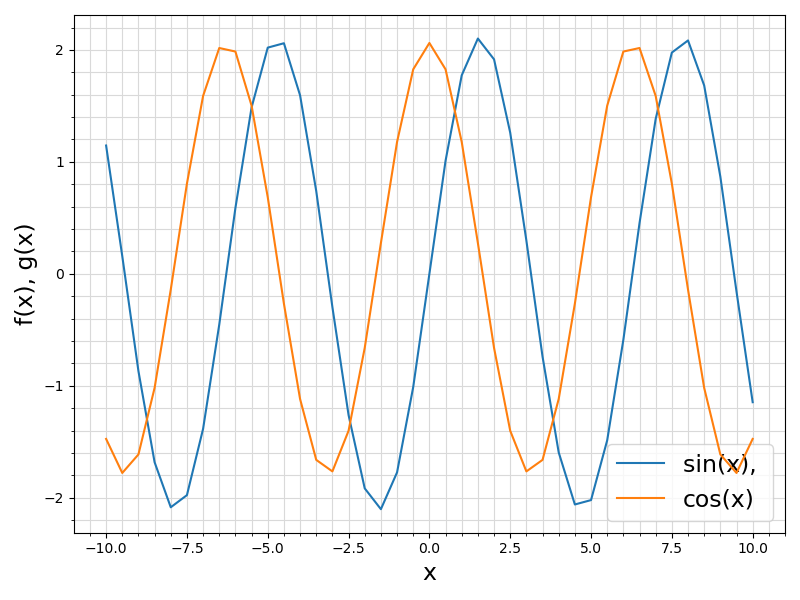
Final Remarks ⛓
If you believe this algorithm or its documentation can be improved, we appreciate your contribution and help to edit this page's documentation and source file on GitHub.
For details on the naming abbreviations, see this page.
For details on the naming conventions, see this page.
This software is distributed under the MIT license with additional terms outlined below.
This software is available to the public under a highly permissive license.
Help us justify its continued development and maintenance by acknowledging its benefit to society, distributing it, and contributing to it.
Definition at line 1051 of file pm_sampleCCF.F90.